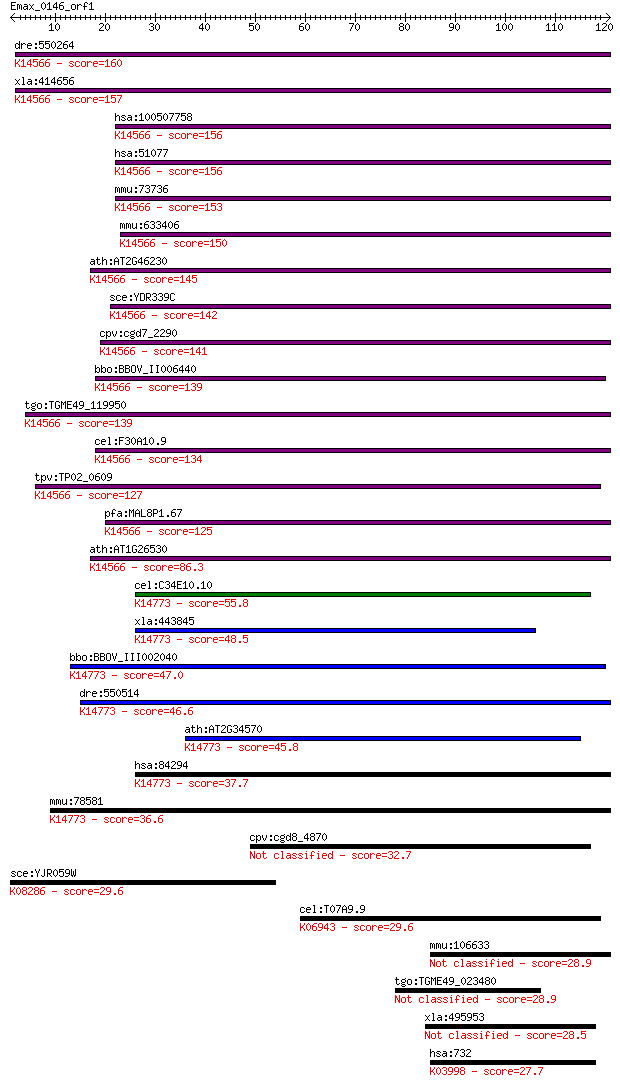
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0146_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
dre:550264 fcf1, zgc:110288; FCF1 small subunit (SSU) processo... 160 8e-40
xla:414656 fcf1, MGC81107; FCF1 small subunit (SSU) processome... 157 7e-39
hsa:100507758 rRNA-processing protein FCF1 homolog; K14566 U3 ... 156 1e-38
hsa:51077 FCF1, Bka, C14orf111, MGC99629; FCF1 small subunit (... 156 1e-38
mmu:73736 Fcf1, 1110008B24Rik; FCF1 small subunit (SSU) proces... 153 1e-37
mmu:633406 rRNA-processing protein FCF1 homolog; K14566 U3 sma... 150 1e-36
ath:AT2G46230 hypothetical protein; K14566 U3 small nucleolar ... 145 3e-35
sce:YDR339C FCF1, UTP24; Fcf1p; K14566 U3 small nucleolar RNA-... 142 2e-34
cpv:cgd7_2290 hypothetical protein ; K14566 U3 small nucleolar... 141 4e-34
bbo:BBOV_II006440 18.m06530; hypothetical protein; K14566 U3 s... 139 2e-33
tgo:TGME49_119950 rRNA-processing protein FCF1, putative ; K14... 139 3e-33
cel:F30A10.9 hypothetical protein; K14566 U3 small nucleolar R... 134 9e-32
tpv:TP02_0609 hypothetical protein; K14566 U3 small nucleolar ... 127 7e-30
pfa:MAL8P1.67 UTP24; nucleolar preribosomal assembly protein; ... 125 4e-29
ath:AT1G26530 hypothetical protein; K14566 U3 small nucleolar ... 86.3 2e-17
cel:C34E10.10 hypothetical protein; K14773 U3 small nucleolar ... 55.8 3e-08
xla:443845 utp23, MGC83220; UTP23, small subunit (SSU) process... 48.5 5e-06
bbo:BBOV_III002040 17.m07198; hypothetical protein; K14773 U3 ... 47.0 1e-05
dre:550514 utp23, fc04b12, wu:fc04b12, zgc:110214; UTP23, smal... 46.6 2e-05
ath:AT2G34570 MEE21; MEE21 (maternal effect embryo arrest 21);... 45.8 3e-05
hsa:84294 UTP23, C8orf53, MGC14595; UTP23, small subunit (SSU)... 37.7 0.009
mmu:78581 Utp23, 1700010I21Rik, AI662478, BB238373, D530033C11... 36.6 0.021
cpv:cgd8_4870 tRNA synthetase class II 32.7 0.28
sce:YJR059W PTK2, STK2; Ptk2p (EC:2.7.11.1); K08286 protein-se... 29.6 2.2
cel:T07A9.9 hypothetical protein; K06943 nucleolar GTP-binding... 29.6 2.3
mmu:106633 Ift140, AI661311, MGC170632, Tce5, Wdtc2, mKIAA0590... 28.9 4.0
tgo:TGME49_023480 thrombospondin type 1 domain-containing prot... 28.9 4.1
xla:495953 c11orf70; chromosome 11 open reading frame 70 28.5
hsa:732 C8B, MGC163447; complement component 8, beta polypepti... 27.7 8.6
> dre:550264 fcf1, zgc:110288; FCF1 small subunit (SSU) processome
component homolog (S. cerevisiae); K14566 U3 small nucleolar
RNA-associated protein 24
Length=198
Score = 160 bits (406), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 75/129 (58%), Positives = 99/129 (76%), Gaps = 10/129 (7%)
Query 2 REREKERAKKEQAK---------HHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKN 52
R +EK+RAK ++ K A S +FF+YN QLGPP+ ++ DTNF+NFSIK
Sbjct 23 RIKEKDRAKTQKKKKEDPSAIKEQEIAKYPSCLFFQYNTQLGPPYYILVDTNFINFSIKA 82
Query 53 KLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYAD 111
KLDV Q+MMDCL AKC+P +TDC++AEL+KLG KYR+AL+I K+ +F+RLPC+HKGTYAD
Sbjct 83 KLDVVQSMMDCLYAKCVPCITDCVMAELEKLGMKYRVALRIAKDPRFDRLPCSHKGTYAD 142
Query 112 DCIVNRVSQ 120
DC+V RV+Q
Sbjct 143 DCLVQRVTQ 151
> xla:414656 fcf1, MGC81107; FCF1 small subunit (SSU) processome
component homolog; K14566 U3 small nucleolar RNA-associated
protein 24
Length=197
Score = 157 bits (398), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 73/128 (57%), Positives = 96/128 (75%), Gaps = 9/128 (7%)
Query 2 REREKERAKKEQAKHHAATV--------SSTMFFRYNLQLGPPFRVICDTNFVNFSIKNK 53
R +EK+RAK + K + S +FF+YN LGPP+ ++ DTNF+NFSIK K
Sbjct 23 RIKEKDRAKIQPKKEDPTAIKEREIPQLPSCLFFQYNTNLGPPYYILVDTNFINFSIKAK 82
Query 54 LDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADD 112
LD+ Q+MMDCL AKC+P +TDC++AEL+KLGQKYR+AL+I K+ FERLPC+H GTYADD
Sbjct 83 LDLVQSMMDCLYAKCVPCITDCVMAELEKLGQKYRVALRIAKDPSFERLPCSHPGTYADD 142
Query 113 CIVNRVSQ 120
C+V RV+Q
Sbjct 143 CLVQRVTQ 150
> hsa:100507758 rRNA-processing protein FCF1 homolog; K14566 U3
small nucleolar RNA-associated protein 24
Length=198
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 68/100 (68%), Positives = 87/100 (87%), Gaps = 1/100 (1%)
Query 22 SSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQ 81
S +FF+YN QLGPP+ ++ DTNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++
Sbjct 52 PSCLFFQYNTQLGPPYHILVDTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIE 111
Query 82 KLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
KLGQKYR+AL+I K+ +FERLPC HKGTYADDC+V RV+Q
Sbjct 112 KLGQKYRVALRIAKDPRFERLPCTHKGTYADDCLVQRVTQ 151
> hsa:51077 FCF1, Bka, C14orf111, MGC99629; FCF1 small subunit
(SSU) processome component homolog (S. cerevisiae); K14566
U3 small nucleolar RNA-associated protein 24
Length=198
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 68/100 (68%), Positives = 87/100 (87%), Gaps = 1/100 (1%)
Query 22 SSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQ 81
S +FF+YN QLGPP+ ++ DTNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++
Sbjct 52 PSCLFFQYNTQLGPPYHILVDTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIE 111
Query 82 KLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
KLGQKYR+AL+I K+ +FERLPC HKGTYADDC+V RV+Q
Sbjct 112 KLGQKYRVALRIAKDPRFERLPCTHKGTYADDCLVQRVTQ 151
> mmu:73736 Fcf1, 1110008B24Rik; FCF1 small subunit (SSU) processome
component homolog (S. cerevisiae); K14566 U3 small nucleolar
RNA-associated protein 24
Length=198
Score = 153 bits (387), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 66/100 (66%), Positives = 87/100 (87%), Gaps = 1/100 (1%)
Query 22 SSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQ 81
S +FF+YN QLGPP+ ++ DTNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++
Sbjct 52 PSCLFFQYNTQLGPPYHILVDTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIE 111
Query 82 KLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
KLGQK+R+AL+I K+ +F+RLPC HKGTYADDC+V RV+Q
Sbjct 112 KLGQKFRVALRIAKDPRFDRLPCTHKGTYADDCLVQRVTQ 151
> mmu:633406 rRNA-processing protein FCF1 homolog; K14566 U3 small
nucleolar RNA-associated protein 24
Length=198
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 65/99 (65%), Positives = 85/99 (85%), Gaps = 1/99 (1%)
Query 23 STMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQK 82
S +FF+YN QLGPP+ ++ TNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++K
Sbjct 53 SCLFFQYNTQLGPPYHILVRTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIEK 112
Query 83 LGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
LGQ YR+AL+I K+ +F+RLPC HKGTYADDC+V RV+Q
Sbjct 113 LGQTYRVALRIAKDPRFDRLPCTHKGTYADDCLVQRVTQ 151
> ath:AT2G46230 hypothetical protein; K14566 U3 small nucleolar
RNA-associated protein 24
Length=181
Score = 145 bits (366), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 64/105 (60%), Positives = 85/105 (80%), Gaps = 1/105 (0%)
Query 17 HAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCI 76
+ +V + +FF YN L PP+RV+ DTNF+NFSI+NK+D+ + M DCL A C P +TDC+
Sbjct 43 NVPSVPAGLFFSYNSTLVPPYRVLVDTNFINFSIQNKIDLEKGMRDCLYANCTPCITDCV 102
Query 77 IAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
+AEL+KLGQKYR+AL+I K+ FERLPC HKGTYADDC+V+RV+Q
Sbjct 103 MAELEKLGQKYRVALRIAKDPHFERLPCIHKGTYADDCLVDRVTQ 147
> sce:YDR339C FCF1, UTP24; Fcf1p; K14566 U3 small nucleolar RNA-associated
protein 24
Length=189
Score = 142 bits (358), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 61/101 (60%), Positives = 86/101 (85%), Gaps = 1/101 (0%)
Query 21 VSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAEL 80
VSS +FF+YN + PP++V+ DTNF+NFSI+ K+D+ + MMDCLLAKC P++TDC++AEL
Sbjct 47 VSSALFFQYNQAIKPPYQVLIDTNFINFSIQKKVDIVRGMMDCLLAKCNPLITDCVMAEL 106
Query 81 QKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
+KLG KYR+ALK+ ++ + +RL C+HKGTYADDC+V+RV Q
Sbjct 107 EKLGPKYRIALKLARDPRIKRLSCSHKGTYADDCLVHRVLQ 147
> cpv:cgd7_2290 hypothetical protein ; K14566 U3 small nucleolar
RNA-associated protein 24
Length=156
Score = 141 bits (356), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 83/103 (80%), Gaps = 1/103 (0%)
Query 19 ATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIA 78
V S++FF YN LGPPF ++ DTNF+NFSI++KLD+ + MDCL+AKCIP +TDC+I
Sbjct 5 PNVPSSLFFNYNNNLGPPFHILLDTNFINFSIQHKLDIFKGTMDCLVAKCIPCITDCVIG 64
Query 79 ELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
EL+K+G +Y LALK+ ++ +F+RL C HKGTYADDCIV RV++
Sbjct 65 ELEKMGHRYNLALKLARDPRFKRLHCLHKGTYADDCIVQRVTE 107
> bbo:BBOV_II006440 18.m06530; hypothetical protein; K14566 U3
small nucleolar RNA-associated protein 24
Length=190
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 66/103 (64%), Positives = 81/103 (78%), Gaps = 1/103 (0%)
Query 18 AATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCII 77
A V S MFF YN L PP++VI DTNFVN SI+NKLD+ + M+DCL+AKCI VTDC+I
Sbjct 42 APKVHSGMFFNYNENLVPPYQVIVDTNFVNASIQNKLDLHKGMLDCLIAKCIVCVTDCVI 101
Query 78 AELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVS 119
AEL+KLG +YRLAL++VK+ + RL C H GTYADDCIV RV+
Sbjct 102 AELEKLGHRYRLALQLVKDPRVSRLTCVHTGTYADDCIVERVT 144
> tgo:TGME49_119950 rRNA-processing protein FCF1, putative ; K14566
U3 small nucleolar RNA-associated protein 24
Length=201
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/118 (53%), Positives = 88/118 (74%), Gaps = 1/118 (0%)
Query 4 REKERAKKEQAKHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDC 63
RE ++ ++E+ +S+MFF YN L PP++VI DTNFVN +++ K DV + +MDC
Sbjct 34 REAKKKQREETIREHVQANSSMFFLYNTNLVPPYQVIVDTNFVNAAVQIKTDVIKGLMDC 93
Query 64 LLAKCIPMVTDCIIAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
L+AKCIP +TDC +AEL+KLG +YRLAL + K+ +F+RL C H GTYADDCIV RV++
Sbjct 94 LVAKCIPCITDCAVAELEKLGHRYRLALALAKDRRFKRLTCCHPGTYADDCIVRRVTE 151
> cel:F30A10.9 hypothetical protein; K14566 U3 small nucleolar
RNA-associated protein 24
Length=196
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 60/104 (57%), Positives = 83/104 (79%), Gaps = 2/104 (1%)
Query 18 AATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCII 77
A +SS MF +YN QLGPPF VI DTNFVNF++KN++D+ Q MDCL AK I TDC++
Sbjct 47 APKISSAMFMKYNTQLGPPFHVIVDTNFVNFAVKNRIDMFQGFMDCLFAKTIVYATDCVL 106
Query 78 AELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
AEL+K+ +++++ALK++K+ + +RL C HKGTYADDC+V RV+Q
Sbjct 107 AELEKV-RRFKIALKVLKDPRVQRLKCEHKGTYADDCLVQRVTQ 149
> tpv:TP02_0609 hypothetical protein; K14566 U3 small nucleolar
RNA-associated protein 24
Length=201
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 70/127 (55%), Positives = 85/127 (66%), Gaps = 14/127 (11%)
Query 6 KERAKKEQAKH--HAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDC 63
KE KKE+ + V S MFF YN L PPF+VI DTNFVN SI+NKLD+ ++MMD
Sbjct 28 KEIEKKEEGPKLTQSNVVHSGMFFNYNENLVPPFQVIVDTNFVNSSIQNKLDLHKSMMDL 87
Query 64 LLAK-----------CIPMVTDCIIAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYAD 111
LL+K I VTDCII EL+KLG +YRLAL++VK+ + +RL C HKGTY D
Sbjct 88 LLSKFTAAEIEEIFLGIICVTDCIIGELEKLGHRYRLALQLVKDPRIKRLKCTHKGTYVD 147
Query 112 DCIVNRV 118
DCIV RV
Sbjct 148 DCIVERV 154
> pfa:MAL8P1.67 UTP24; nucleolar preribosomal assembly protein;
K14566 U3 small nucleolar RNA-associated protein 24
Length=198
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 77/102 (75%), Gaps = 1/102 (0%)
Query 20 TVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAE 79
+ S +FF YN L PP+ +I DTNF+N SI+ K+D+ + + LLAKC VTDC+I E
Sbjct 52 VIDSNLFFNYNENLCPPYNIILDTNFINSSIQYKIDIIKGCSEVLLAKCNIFVTDCVIGE 111
Query 80 LQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
++KLGQ+Y L LK++K+ +F+RL C HKGTYADDCIVNRV++
Sbjct 112 MEKLGQRYSLGLKLIKDPRFKRLTCNHKGTYADDCIVNRVTE 153
> ath:AT1G26530 hypothetical protein; K14566 U3 small nucleolar
RNA-associated protein 24
Length=178
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 45/104 (43%), Positives = 63/104 (60%), Gaps = 17/104 (16%)
Query 17 HAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCI 76
+ +V S +FF +N L PP+RV+ DTNF+NFSI+NK+D+ + MM
Sbjct 43 NVPSVPSGLFFSHNSSLVPPYRVLVDTNFINFSIQNKIDLEKGMM-------------VF 89
Query 77 IAELQKLGQKYRLALKIVKEQFERLPCAHKGTYADDCIVNRVSQ 120
+ ++ L Y L K +FERLPC KGTYADDC+V+RV+Q
Sbjct 90 VRKMHSL--YYGLIAK--DPRFERLPCVLKGTYADDCLVDRVTQ 129
> cel:C34E10.10 hypothetical protein; K14773 U3 small nucleolar
RNA-associated protein 23
Length=232
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query 26 FFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQ 85
F++YN + P+RV+ D F N +++ KL++ + + L + M T+C++ EL+K G
Sbjct 15 FYKYNYKFVAPYRVLVDGTFCNAALQEKLNLAEQIPKYLTEETHLMTTNCVLKELEKFGP 74
Query 86 KYRLALKIVKEQFERLPCAHKGT-YADDCIVN 116
A I K QFE C H A DC+ +
Sbjct 75 LLYGAFVIAK-QFEIAECTHSTPRAASDCLAH 105
> xla:443845 utp23, MGC83220; UTP23, small subunit (SSU) processome
component, homolog; K14773 U3 small nucleolar RNA-associated
protein 23
Length=243
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Query 26 FFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQ 85
F+R+N + P++V+ D F ++KNK+ + + + L+ + + C+I EL+ LG+
Sbjct 15 FYRFNFGVREPYQVLLDGTFCQAALKNKIQIKEQLPKYLMGEVQLCTSHCVIKELESLGK 74
Query 86 KYRLALKIVKEQFERLPCAH 105
+ A K++ ++F+ C+H
Sbjct 75 ELYGA-KLIAQRFQVRSCSH 93
> bbo:BBOV_III002040 17.m07198; hypothetical protein; K14773 U3
small nucleolar RNA-associated protein 23
Length=256
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 58/111 (52%), Gaps = 5/111 (4%)
Query 13 QAKHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMV 72
+A H + F+R + L P+R++ D +F ++K+K+++ + + + L V
Sbjct 2 KANKHKRVKRTLDFYRQLVNLTVPYRILVDGSFAFAALKHKVNIKEHLTELLGDTTHTYV 61
Query 73 TDCIIAELQKLGQKYRLALKIVKEQFERLPCAH----KGTYADDCIVNRVS 119
T CII EL+ +G++ A+ +K + +RL C H K + CI + VS
Sbjct 62 TSCIIDELRGMGEEMSGAVLALK-RCQRLRCNHQPSDKAPNSRRCITSAVS 111
> dre:550514 utp23, fc04b12, wu:fc04b12, zgc:110214; UTP23, small
subunit (SSU) processome component, homolog (yeast); K14773
U3 small nucleolar RNA-associated protein 23
Length=249
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 57/108 (52%), Gaps = 6/108 (5%)
Query 15 KHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTD 74
KH T+S F++YN PF+++ D F ++KNK+ + + + L+ + T+
Sbjct 7 KHAKKTIS---FYKYNFSFREPFQILIDGTFCQAALKNKIQIKEQLPKYLMGEIQLCTTN 63
Query 75 CIIAELQKLGQKYRLALKIVKEQFERLPCAH--KGTYADDCIVNRVSQ 120
C + EL+ L + A K++ ++F+ C H A +C+++ +++
Sbjct 64 CALKELESLAKDLYGA-KLILQRFQIRKCKHMKDPVPASECLLSMLAE 110
> ath:AT2G34570 MEE21; MEE21 (maternal effect embryo arrest 21);
K14773 U3 small nucleolar RNA-associated protein 23
Length=281
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 36 PFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPM-VTDCIIAELQKLGQKYRLALKIV 94
P++V+CD FV+ + N++ + LL + + T C+IAEL+KLG+ + +L+
Sbjct 25 PYKVLCDGTFVHHLVTNEITPADTAVSELLGGPVKLFTTRCVIAELEKLGKDFAESLEAA 84
Query 95 KEQFERLPCAH-KGTYADDCI 114
+ C H + AD+C+
Sbjct 85 -QTLNTATCEHEEAKTADECL 104
> hsa:84294 UTP23, C8orf53, MGC14595; UTP23, small subunit (SSU)
processome component, homolog (yeast); K14773 U3 small nucleolar
RNA-associated protein 23
Length=249
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/97 (20%), Positives = 49/97 (50%), Gaps = 3/97 (3%)
Query 26 FFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQ 85
FFR N + P++++ D F +++ ++ + + + L+ + T C++ EL+ LG+
Sbjct 15 FFRNNFGVREPYQILLDGTFCQAALRGRIQLREQLPRYLMGETQLCTTRCVLKELETLGK 74
Query 86 KYRLALKIVKEQFERLPCAH--KGTYADDCIVNRVSQ 120
A K++ ++ + C H +C+++ V +
Sbjct 75 DLYGA-KLIAQKCQVRNCPHFKNAVSGSECLLSMVEE 110
> mmu:78581 Utp23, 1700010I21Rik, AI662478, BB238373, D530033C11Rik;
UTP23, small subunit (SSU) processome component, homolog
(yeast); K14773 U3 small nucleolar RNA-associated protein
23
Length=249
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 23/114 (20%), Positives = 55/114 (48%), Gaps = 9/114 (7%)
Query 9 AKKEQAKHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKC 68
+++ AK H FFR N + P++++ D F +++ ++ + + L+ +
Sbjct 4 TRQKHAKKHLG------FFRNNFGVREPYQILLDGTFCQAALRGRIQLRDQLPRYLMGET 57
Query 69 IPMVTDCIIAELQKLGQKYRLALKIVKEQFERLPCAH--KGTYADDCIVNRVSQ 120
T C++ EL+ LG++ A K++ ++ + C H +C+++ V +
Sbjct 58 QLCTTRCVLKELETLGKELYGA-KLIAQKCQVRNCPHFKSPVSGSECLLSMVDE 110
> cpv:cgd8_4870 tRNA synthetase class II
Length=216
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 7/73 (9%)
Query 49 SIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQKYR-----LALKIVKEQFERLPC 103
++KNK+ + + L K P++TDCI E++ L K++ + R C
Sbjct 3 ALKNKIHIKEQFPKILNGKTTPIITDCIYKEIEMLNNLESKKADFSGAKLIARGYFRHKC 62
Query 104 AHKGTYADDCIVN 116
H TY ++ I N
Sbjct 63 GH--TYCNENISN 73
> sce:YJR059W PTK2, STK2; Ptk2p (EC:2.7.11.1); K08286 protein-serine/threonine
kinase [EC:2.7.11.-]
Length=818
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 1 DREREKERAK-KEQAKHHAA------TVSSTMFFRYNLQLGP----PFRVICDTNFVNFS 49
DRE+EKERA+ KE+ HHA +++S F N+ P P DT F +
Sbjct 145 DREKEKERARNKERNTHHAGLPQRSNSMASHHFPNENIVYNPYGISPNHARPDTAFADTL 204
Query 50 IKNK 53
NK
Sbjct 205 NTNK 208
> cel:T07A9.9 hypothetical protein; K06943 nucleolar GTP-binding
protein
Length=681
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 59 AMMDCLLAKCIPMVTDCIIAELQKLGQKYRLALKIVKEQFERLPCAHKGTYADDCIVNRV 118
A++D L + IP++ + + +G + R +++ ++ E A K T +DC++NRV
Sbjct 306 ALLDQLEKEGIPIIETSTLTQEGVMGLRDRACDELLAQRVEAKIQAKKITNVEDCVLNRV 365
> mmu:106633 Ift140, AI661311, MGC170632, Tce5, Wdtc2, mKIAA0590;
intraflagellar transport 140 homolog (Chlamydomonas)
Length=1464
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 85 QKYRLALKIVKEQFERLPCAHKGTYADDCIVNRVSQ 120
++Y++A K ++E +RLP A+ Y D V+ V Q
Sbjct 1391 EEYQMAYKYLEEMRKRLPSANMSYYVDQRTVDTVHQ 1426
> tgo:TGME49_023480 thrombospondin type 1 domain-containing protein
(EC:3.4.24.14 2.7.8.17 3.1.1.2 3.4.24.82)
Length=4491
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 78 AELQKLGQKYRLALKIVKEQFERLPCAHK 106
A ++++G +L LK+ KEQF L CA K
Sbjct 1873 AAVKEMGYSCKLLLKVAKEQFPALGCAAK 1901
> xla:495953 c11orf70; chromosome 11 open reading frame 70
Length=203
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 84 GQKYRLALKIVKEQFERLPCAHKGTYADDCIVNR 117
GQ RL K+ K E +PC+H DC+ +
Sbjct 21 GQWVRLGTKVKKVDVEEIPCSHLSMSIFDCLYSE 54
> hsa:732 C8B, MGC163447; complement component 8, beta polypeptide;
K03998 complement component 8 subunit beta
Length=591
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 85 QKYRLALKIVKEQFERLPCAHKGTYADDCIVNR 117
++YR A + QF PC +DC+ NR
Sbjct 82 KRYRYAYLLQPSQFHGEPCNFSDKEVEDCVTNR 114
Lambda K H
0.326 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40