bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0130_orf3
Length=560
Score E
Sequences producing significant alignments: (Bits) Value
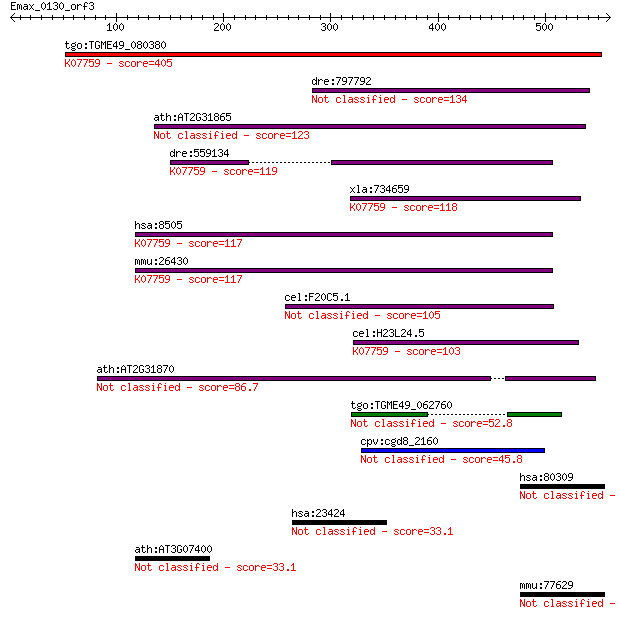
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 405 3e-112
dre:797792 si:dkey-259k14.2 134 7e-31
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 123 2e-27
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 119 2e-26
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 118 6e-26
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 117 1e-25
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 117 1e-25
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 105 5e-22
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 103 3e-21
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 86.7 2e-16
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 52.8 4e-06
cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydro... 45.8 5e-04
hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132... 35.0 0.69
hsa:23424 TDRD7, KIAA1529, PCTAIRE2BP, RP11-508D10.1, TRAP; tu... 33.1 2.8
ath:AT3G07400 lipase class 3 family protein 33.1 3.1
mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIA... 32.7 4.3
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 405 bits (1040), Expect = 3e-112, Method: Compositional matrix adjust.
Identities = 210/513 (40%), Positives = 304/513 (59%), Gaps = 17/513 (3%)
Query 52 DTPYAVLRPAEESHQRRIQDILRRIEDGGVPSTAAQFEALFVHLLREAGNYRGGGQ---L 108
D P A+LR E + + IL + G P+ Q +L LL+ +GN R L
Sbjct 43 DNPCAILRTPVEEDWGKTKYILNFVASGFGPADPQQLASLQKRLLKLSGNIRTEKNKRIL 102
Query 109 QIGRLNSYLAQDEEFSRTTLPFLASVAKHISDLFPKGLKHMTRSHPSVHLRRIQVLCLMA 168
Q G LN +F LPF+A++ I +LFP GL+++T +P VHLR+IQV L+A
Sbjct 103 QRGLLNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVFTLIA 162
Query 169 ASFFGIIPQKQRKLLRSWGR---LRLNALDMHHRGFFEREPKLKALLIYFGSMQKTMSGC 225
A+F G+IP QR LL + + N LDM +RGF ER+PK +++LIYF SM++ + C
Sbjct 163 AAFLGVIPHNQRALLAHHQKKFVMNQNKLDMFYRGFMERKPKFQSVLIYFASMRERLGNC 222
Query 226 WQQMLKDNDFTQAKCTATCLCQEITFEGRTRKISPADEIISFYLHTPEATTFEAPEGE-Q 284
W +M+K A CT TC+C + + I+P DE+I FY + + F +G +
Sbjct 223 WNEMVKKGFVDMAPCTGTCICSTLE---ESVSIAPTDELIGFYRQSDKVADFVWSDGSVK 279
Query 285 VSRSEVDVSKIISQPLPLSGLTVDVHGDITKSDGDLQVDFADMYVGGLSMWAGHVAQEEL 344
+ + V + I L G V GDIT SDGDLQVDFAD Y+GGLSM+ G++AQEEL
Sbjct 280 GTSASVSIDDIAKSTKALGGFEVFEEGDITTSDGDLQVDFADKYLGGLSMFEGYIAQEEL 339
Query 345 IFALRPELHVIMLFREALRDDEAVVVKGAESFVLYSGYEGTFQVFPPV-----VPAGSHW 399
IF + PE+ +MLF + ++ +EAVVVKG E FV GYE TFQ+ +P+ H+
Sbjct 340 IFIIYPEMLCVMLFSDIMKPNEAVVVKGVERFVFSQGYEWTFQITAAAGDWTELPSNKHF 399
Query 400 SVHGLAPLDSMGRRETTVVAIDAIVVKNEREQYSIQSMKREILKAALGFQGDPFEDVIHE 459
SV G+ PLDS+GRR +V IDA+ +QYS + RE++KAA+GF+GDP+E ++
Sbjct 400 SVQGVVPLDSLGRRNVAIVGIDAVQFHEPNKQYSPVMVNRELMKAAVGFKGDPYELIVSG 459
Query 460 TRAGVATGKWGCVIFGGDNQLKSLLQWIAATAAGREMHYKAFGDATLAHMEGTLSAIRHR 519
+R +ATG WGC +F GD QLK+L+QW+AA+ AGR M + F + ++ + +S +R
Sbjct 460 SRQPIATGLWGCGVFNGDAQLKTLIQWLAASYAGRSMKFYTFSNKSVDGLGLVISKLRQS 519
Query 520 FPTTKDLFEAIVQCIQQREETPIGNWSLWQSLL 552
+ T +L++AI + +R WSLW+ LL
Sbjct 520 YATVGNLYKAIQNALSRRSGRL--GWSLWKELL 550
> dre:797792 si:dkey-259k14.2
Length=609
Score = 134 bits (338), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 93/260 (35%), Positives = 132/260 (50%), Gaps = 20/260 (7%)
Query 283 EQVSRSEVDVSKIISQPLPLSGLTVDVHGDITKS-DGDLQVDFADMYVGGLSMWAGHVAQ 341
E+VS ++ K S+ L L V G I K G LQVDFA ++GG + +G V Q
Sbjct 347 ERVSVPASELPKWKSEKKLLKNLHVSADGSIEKEGTGMLQVDFASKFIGGGVLKSGLV-Q 405
Query 342 EELIFALRPELHVIMLFREALRDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPAGSHWSV 401
EE++F + PEL + LF E L D E V + G + + L SGY +F P +
Sbjct 406 EEILFLMSPELILARLFTEKLDDHECVRITGPQMYSLTSGYSRSFSWTGPYMDRTKR--- 462
Query 402 HGLAPLDSMGRRETTVVAIDAIVVKNEREQYSIQSMKREILKAALGFQGDPFEDVIHETR 461
D RR +VAIDA+ KN EQYS +++ RE+ KA +GF G P +
Sbjct 463 ------DVWKRRFRQIVAIDALDFKNPLEQYSRENITRELNKAFVGFCGQP--------K 508
Query 462 AGVATGKWGCVIFGGDNQLKSLLQWIAATAAGREMHYKAFGDATLAH-MEGTLSAIRHRF 520
+ATG WGC F GD +LK+LLQ +AA R++ Y FG+ LA+ ++ + R
Sbjct 509 TAIATGNWGCGAFRGDPKLKALLQLMAAAVVDRDVAYFTFGNTHLANELQKMHDILTQRK 568
Query 521 PTTKDLFEAIVQCIQQREET 540
T L+E + + E T
Sbjct 569 VTVGKLYELLKDYCKHYERT 588
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 123 bits (308), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 120/441 (27%), Positives = 192/441 (43%), Gaps = 94/441 (21%)
Query 135 AKHISDLFPKGLKHMTRSHPSVHLRRIQVLC-LMAASFFGIIPQKQRKLLRSWGRLRLNA 193
A H+ D GL+ + + L +++ L+A SFF + P+ R L G + +
Sbjct 119 ADHVLDGVKSGLRLLGPQEAGIVLLSQELIAALLACSFFCLFPEVDRSLKNLQG-INFSG 177
Query 194 LDM--HHRGFFEREPKLKALLIYFGSMQKTMSGCWQQMLKDNDFTQAKCTATCLCQEITF 251
L + R ++E K+K L+ YFG + + M + ++F
Sbjct 178 LFSFPYMRHCTKQENKIKCLIHYFGRICRWMPTGF----------------------VSF 215
Query 252 EGRTRKISPADEIISFYLHTPEATTFEAPEGEQVSRSEVDVSKIISQPLPLSGLTVDVHG 311
E RKI P + + P ++ P+ + + S PL + + G
Sbjct 216 E---RKILPLE-------YHPHFVSY--PKADSWANSVT----------PLCSIEIHTSG 253
Query 312 DITKSDGD-LQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVV 370
I + L+VDFAD Y GGL++ + QEE+ F + PEL M+F + +EA+ +
Sbjct 254 AIEDQPCEALEVDFADEYFGGLTL-SYDTLQEEIRFVINPELIAGMIFLPRMDANEAIEI 312
Query 371 KGAESFVLYSGYEGTFQVFPPVVPAGSHWSVHGLAPLDSMGRRETTVVAIDAIVVKNERE 430
G E F Y+GY +FQ AG + LD RR+T V+AIDA+
Sbjct 313 VGVERFSGYTGYGPSFQY------AGDYTDNKD---LDIFRRRKTRVIAIDAMPDPG-MG 362
Query 431 QYSIQSMKREILKAALGF----------QGDPFEDVIH---------------------- 458
QY + ++ RE+ KA G+ + DP H
Sbjct 363 QYKLDALIREVNKAFSGYMHQCKYNIDVKHDPEASSSHVPLTSDSASQVIESSHRWCIDH 422
Query 459 -ETRAGVATGKWGCVIFGGDNQLKSLLQWIAATAAGRE-MHYKAFGDATLAHMEGTLSAI 516
E + GVATG WGC +FGGD +LK +LQW+A + +GR M Y FG L ++ + +
Sbjct 423 EEKKIGVATGNWGCGVFGGDPELKIMLQWLAISQSGRPFMSYYTFGLQALQNLNQVIEMV 482
Query 517 RHRFPTTKDLFEAIVQCIQQR 537
+ T DL++ +V+ +R
Sbjct 483 ALQEMTVGDLWKKLVEYSSER 503
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 119 bits (299), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 80/209 (38%), Positives = 112/209 (53%), Gaps = 18/209 (8%)
Query 301 PLSGLTVDVHGDITKSD-GDLQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFR 359
PLS L + G I G LQVDFA+ + G + + QEE+ F + PEL V LF
Sbjct 508 PLSHLHITCKGTIEDQGYGMLQVDFANR-MVGGGVTGLGLVQEEIRFLINPELIVSRLFT 566
Query 360 EALRDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPAGSHWSVH--GLAPLDSMGRRETTV 417
E L +E +++ G E + YSGY +F+ W + P D RR T +
Sbjct 567 EVLDHNECLIITGTEQYSKYSGYAESFK-----------WEDNHKDKIPRDGWQRRCTEI 615
Query 418 VAIDAIVVKNEREQYSIQSMKREILKAALGFQGDPFEDVIHETRAGVATGKWGCVIFGGD 477
VA+DA+ +N +Q+ + M RE+ KA GF P + ++ + VATG WGC FGGD
Sbjct 616 VAMDALHYRNFMDQFQPEKMTRELNKAYCGFMR-PGVNPLN--LSAVATGNWGCGAFGGD 672
Query 478 NQLKSLLQWIAATAAGREMHYKAFGDATL 506
+LK+LLQ +AA AGR++ Y FGD L
Sbjct 673 TRLKALLQLMAAAEAGRDVAYFTFGDEAL 701
Score = 34.3 bits (77), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 150 TRSHPSVHLRRIQVLCLMAASFFGIIPQKQRKLLRSWGRLRLNALDMHHRGFFEREPKLK 209
T+ + S+ + + Q+ CL+A +FF P++ + +N + + KLK
Sbjct 412 TKMNQSLTMSQEQIACLLANAFFCTFPRRNSRKSEYANYPEINFYRLFEGSSQRKIEKLK 471
Query 210 ALLIYFGSMQKTM 222
LL YF + ++M
Sbjct 472 TLLCYFRRVTESM 484
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 118 bits (296), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 80/218 (36%), Positives = 119/218 (54%), Gaps = 18/218 (8%)
Query 318 GDLQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESFV 377
G LQVDFA+ +VGG G + QEE+ F + PEL V LF E L +E +++ GAE +
Sbjct 529 GMLQVDFANRFVGGGVT-GGGLVQEEIRFLINPELIVSRLFTEVLDSNECLIITGAEQYS 587
Query 378 LYSGYEGTFQVFPPVVPAGSHWS-VH-GLAPLDSMGRRETTVVAIDAIVVKNEREQYSIQ 435
Y+GY T++ W+ VH +P D RR T +VAIDA + +Q+ +
Sbjct 588 EYTGYSETYK-----------WACVHEDESPRDEWQRRTTEIVAIDAFHFRRPIDQFVPE 636
Query 436 SMKREILKAALGFQGDPFEDVIHETRAGVATGKWGCVIFGGDNQLKSLLQWIAATAAGRE 495
+KRE+ KA GF +V + + VATG WGC FGGD +LK+L+Q +AA GR+
Sbjct 637 KIKRELNKAFCGFYR---PEVNPQNLSAVATGNWGCGAFGGDPRLKALIQLLAAAEVGRD 693
Query 496 MHYKAFGDATL-AHMEGTLSAIRHRFPTTKDLFEAIVQ 532
+ Y FGD L + S + + T D++ +++
Sbjct 694 LVYFTFGDRELMKDIYLMYSFLTEKNKTVGDIYSMLIE 731
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 117 bits (293), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 118/398 (29%), Positives = 182/398 (45%), Gaps = 83/398 (20%)
Query 118 AQDEEFSRTTLPFLASVAKHISDLF--PKGLKHMTRSHPSVHLRRIQVLCLMAASFFGII 175
A+ + ++ LP + +A + ++ P L +H S+ + + Q+ L+A +FF
Sbjct 584 AEAQHLYQSILPDMVKIALCLPNICTQPIPLLKQKMNH-SITMSQEQIASLLANAFFCTF 642
Query 176 PQKQRKL---LRSWGRLRLNALDMHHRGFFEREP-KLKALLIYFGSMQKTMSGCWQQMLK 231
P++ K+ S+ + N L G R+P KLK L YF +
Sbjct 643 PRRNAKMKSEYSSYPDINFNRL---FEGRSSRKPEKLKTLFCYFRRV------------- 686
Query 232 DNDFTQAKCTATCLCQEITFEGRTRKISPADEIISFYLHTPEATTFEAPEGEQVSRSEVD 291
T+ K T +++F + E + PE E+ +
Sbjct 687 ----TEKKPTG---------------------LVTFTRQSLE----DFPEWERCEK---- 713
Query 292 VSKIISQPLPLSGLTVDVHGDITKS-DGDLQVDFADMYVGGLSMWAGHVAQEELIFALRP 350
PL+ L V G I ++ G LQVDFA+ +VGG AG V QEE+ F + P
Sbjct 714 ---------PLTRLHVTYEGTIEENGQGMLQVDFANRFVGGGVTSAGLV-QEEIRFLINP 763
Query 351 ELHVIMLFREALRDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPAGSHWSV--HGLAPLD 408
EL + LF E L +E +++ G E + Y+GY T++ WS + D
Sbjct 764 ELIISRLFTEVLDHNECLIITGTEQYSEYTGYAETYR-----------WSRSHEDGSERD 812
Query 409 SMGRRETTVVAIDAIVVKNEREQYSIQSMKREILKAALGFQGDPFEDVIHETRAGVATGK 468
RR T +VAIDA+ + +Q+ + M+RE+ KA GF V E + VATG
Sbjct 813 DWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCGFLR---PGVSSENLSAVATGN 869
Query 469 WGCVIFGGDNQLKSLLQWIAATAAGREMHYKAFGDATL 506
WGC FGGD +LK+L+Q +AA AA R++ Y FGD+ L
Sbjct 870 WGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSEL 907
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 117 bits (293), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 118/395 (29%), Positives = 181/395 (45%), Gaps = 77/395 (19%)
Query 118 AQDEEFSRTTLPFLASVAKHISDLF--PKGLKHMTRSHPSVHLRRIQVLCLMAASFFGII 175
A+ + ++ LP + +A + ++ P L +H SV + + Q+ L+A +FF
Sbjct 577 AEAQHLYQSILPDMVKIALCLPNICTQPIPLLKQKMNH-SVTMSQEQIASLLANAFFCTF 635
Query 176 PQKQRKLLRSWGRLRLNALDMHHRGFFEREP-KLKALLIYFGSMQKTMSGCWQQMLKDND 234
P++ K+ + + G R+P KLK L YF +
Sbjct 636 PRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCYFRRV---------------- 679
Query 235 FTQAKCTATCLCQEITFEGRTRKISPADEIISFYLHTPEATTFEAPEGEQVSRSEVDVSK 294
T+ K T +++F + E + PE E+ +
Sbjct 680 -TEKKPTG---------------------LVTFTRQSLE----DFPEWERCEK------- 706
Query 295 IISQPLPLSGLTVDVHGDIT-KSDGDLQVDFADMYVGGLSMWAGHVAQEELIFALRPELH 353
PL+ L V G I G LQVDFA+ +VGG AG V QEE+ F + PEL
Sbjct 707 ------PLTRLHVTYEGTIEGNGRGMLQVDFANRFVGGGVTGAGLV-QEEIRFLINPELI 759
Query 354 VIMLFREALRDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPAGSHWSVHGLAPLDSMGRR 413
V LF E L +E +++ G E + Y+GY T++ A SH + D RR
Sbjct 760 VSRLFTEVLDHNECLIITGTEQYSEYTGYAETYRW------ARSH---EDGSEKDDWQRR 810
Query 414 ETTVVAIDAIVVKNEREQYSIQSMKREILKAALGF--QGDPFEDVIHETRAGVATGKWGC 471
T +VAIDA+ + +Q+ + ++RE+ KA GF G P E++ + VATG WGC
Sbjct 811 CTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCGFLRPGVPSENL-----SAVATGNWGC 865
Query 472 VIFGGDNQLKSLLQWIAATAAGREMHYKAFGDATL 506
FGGD +LK+L+Q +AA AA R++ Y FGD+ L
Sbjct 866 GAFGGDARLKALIQILAAAAAERDVVYFTFGDSEL 900
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 105 bits (262), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 83/257 (32%), Positives = 124/257 (48%), Gaps = 30/257 (11%)
Query 258 ISPADEIISFYLHTPEATTFEAPEGEQVSRSEVDVSKIISQPLPLSGLTVDVHGDITKSD 317
+ P D +SF L + TF ++ RS +V + + L
Sbjct 467 MDPPDGAVSFRLTKMDKDTFNEEWKDKKLRSLPEVEFFDEMLIEDTALCT---------- 516
Query 318 GDLQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESFV 377
QVDFA+ ++GG + G V QEE+ F + PE+ V ML E ++ EA+ + GA F
Sbjct 517 ---QVDFANEHLGGGVLNHGSV-QEEIRFLMCPEMMVGMLLCEKMKQLEAISIVGAYVFS 572
Query 378 LYSGYEGTFQVFPPVVPAGSHWSVHGLAPLDSMGRRETTVVAIDAIVVKNER-----EQY 432
Y+GY T + + + P S + + D GR +AIDAI+ K + EQ
Sbjct 573 SYTGYGHTLK-WAELQPNHSRQNTNEFR--DRFGRLRVETIAIDAILFKGSKLDCQTEQL 629
Query 433 SIQSMKREILKAALGF--QGDPFEDVIHETRAGVATGKWGCVIFGGDNQLKSLLQWIAAT 490
+ ++ RE+ KA++GF QG F T + TG WGC F GD LK ++Q IAA
Sbjct 630 NKANIIREMKKASIGFMSQGPKF------TNIPIVTGWWGCGAFNGDKPLKFIIQVIAAG 683
Query 491 AAGREMHYKAFGDATLA 507
A R +H+ +FG+ LA
Sbjct 684 VADRPLHFCSFGEPELA 700
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 103 bits (256), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 73/218 (33%), Positives = 112/218 (51%), Gaps = 18/218 (8%)
Query 321 QVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESFVLYS 380
Q+DFA+ +GG + G V QEE+ F + PE+ V +L + +D EA+ + GA F Y+
Sbjct 251 QIDFANKRLGGGVLKGGAV-QEEIRFMMCPEMMVAILLNDVTQDLEAISIVGAYVFSSYT 309
Query 381 GYEGTFQVFPPVVPAGSHWSVHGLAPLDSMGRRETTVVAIDAIV-----VKNEREQYSIQ 435
GY T + + + P H + + + D GR +T VAIDA+ ++ Q + +
Sbjct 310 GYSNTLK-WAKITP--KHSAQNNNSFRDQFGRLQTETVAIDAVRNAGTPLECLLNQLTTE 366
Query 436 SMKREILKAALGF--QGDPFEDVIHETRAGVATGKWGCVIFGGDNQLKSLLQWIAATAAG 493
+ RE+ KAA+GF GD F ++ V +G WGC F G+ LK L+Q IA +
Sbjct 367 KLTREVRKAAIGFLSAGDGF------SKIPVVSGWWGCGAFRGNKPLKFLIQVIACGISD 420
Query 494 REMHYKAFGDATLA-HMEGTLSAIRHRFPTTKDLFEAI 530
R + + FGD LA E ++ R+ T LF I
Sbjct 421 RPLQFCTFGDTELAKKCEEMMTLFRNNNVRTGQLFLII 458
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 86.7 bits (213), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 103/372 (27%), Positives = 147/372 (39%), Gaps = 76/372 (20%)
Query 82 PSTAAQFEALFVHLLREAGNYRGGGQLQIGRLNSYLAQDEEFSRTTLPFLASVAKHISDL 141
PS + + LF L+ E + R ++ I L S L Q P L V +D
Sbjct 69 PSASNGYAFLFDELIDEKESKRWFDEI-IPALASLLLQ--------FPSLLEVHFQNADN 119
Query 142 FPKGLKHMTRSHPS-----VHLRRIQVLCLMAASFFGIIPQKQRKLLRSWGRLRLNALDM 196
G+K R S V L + + L+A SFF + P R G L ++
Sbjct 120 IVSGIKTGLRLLNSQQAGIVFLSQELIGALLACSFFCLFPDDNR------GAKHLPVINF 173
Query 197 HHRGFFEREPKLKALLIYFGSMQKTMSGCWQQMLKDNDFTQAKCTATCLCQEITFEGRTR 256
H +L I + Q++ C + C C I R
Sbjct 174 DHL--------FASLYISYSQSQESKIRCIMHYFE----------RFCSCVPIGIVSFER 215
Query 257 KISPADEIISFYLHTPEATTFEAPEGEQVSRSEVDVSKIISQPLPLSGLTVDVHGDITKS 316
KI+ A P+ + S+S+V + GL D +
Sbjct 216 KITAA------------------PDADFWSKSDVSLCAFKVHSF---GLIED------QP 248
Query 317 DGDLQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESF 376
D L+VDFA+ Y+GG S+ G V QEE+ F + PEL MLF + D+EA+ + GAE F
Sbjct 249 DNALEVDFANKYLGGGSLSRGCV-QEEIRFMINPELIAGMLFLPRMDDNEAIEIVGAERF 307
Query 377 VLYSGYEGTFQVFPPVVPAGSHWSVHGLAPLDSMGRRETTVVAIDAIVVKNEREQYSIQS 436
Y+GY +F+ AG + + P RR T +VAIDA+ R I
Sbjct 308 SCYTGYASSFRF------AGEYIDKKAMDPFK---RRRTRIVAIDALCTPKMRHFKDI-C 357
Query 437 MKREILKAALGF 448
+ REI KA GF
Sbjct 358 LLREINKALCGF 369
Score = 57.8 bits (138), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 50/87 (57%), Gaps = 3/87 (3%)
Query 463 GVATGKWGCVIFGGDNQLKSLLQWIAATAAGRE-MHYKAFGDATLAHMEGTLSAIRHRFP 521
GVATG WGC +FGGD +LK+ +QW+AA+ R + Y FG L +++ I
Sbjct 445 GVATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRNLDQVTKWILSHKW 504
Query 522 TTKDLFEAIVQCIQQR--EETPIGNWS 546
T DL+ +++ QR ++T +G +S
Sbjct 505 TVGDLWNMMLEYSAQRLYKQTSVGFFS 531
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 52.8 bits (125), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 34/56 (60%), Gaps = 6/56 (10%)
Query 465 ATGKWGCVIFGGDNQLKSLLQWIAATAAGREMHYKA------FGDATLAHMEGTLS 514
ATG WGC +F GD QLK LLQW+AA+ GR + Y A G A+ + GT++
Sbjct 829 ATGNWGCGVFKGDPQLKFLLQWLAASLVGRRLIYHAHSRPELVGSASRSQRGGTVT 884
Score = 45.8 bits (107), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Query 319 DLQVDFADMYVGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESFVL 378
++ DFA+ ++GG +++ G V QEE+ FA PEL ++ LF++ L +E+ + GA F
Sbjct 479 EIMADFANQWIGGGALYRGCV-QEEIFFATHPELLLLRLFQQRLAINESCAMSGAMQFSR 537
Query 379 YSGYEGTFQVF 389
YSGY +F
Sbjct 538 YSGYADSFTCL 548
> cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydrolases,
some protein kinase A anchoring proteins and baculovirus
HzNV Orf103, possible transmembrane domain within N-terminus
Length=441
Score = 45.8 bits (107), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 42/170 (24%), Positives = 75/170 (44%), Gaps = 12/170 (7%)
Query 329 VGGLSMWAGHVAQEELIFALRPELHVIMLFREALRDDEAVVVKGAESFVLYSGYEGTFQV 388
V G + + QEE++ + PE + F + L D+++V ++G + Y G+
Sbjct 235 VYGSATLQNCMNQEEIVMTVVPETLIGRFFLDDLHDEDSVTIRGVMRYSNYKGFGTDKFT 294
Query 389 FPPVVPAGSHWSVHGLAPLDSMGRRETTVVAIDAIVVKNEREQYSIQSMKREILKAALGF 448
F + + + S+ + + +DA+ + ++++ REI K
Sbjct 295 FQSIKESEMYMSIPRVYAV------------VDALSGGSRFREFTLDYALREINKLISAL 342
Query 449 QGDPFEDVIHETRAGVATGKWGCVIFGGDNQLKSLLQWIAATAAGREMHY 498
D + + R TG WG +FGGDNQ K +LQ IAA R++ Y
Sbjct 343 CDDFYGESEERDRNQFVTGYWGGGVFGGDNQYKFILQLIAACVCNRQLVY 392
> hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132614,
MGC132616, SKIP; SPHK1 interactor, AKAP domain containing
Length=1700
Score = 35.0 bits (79), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 40/81 (49%), Gaps = 5/81 (6%)
Query 477 DNQLKSLLQWIAATAAGREMHYKAFGDATLAHMEGTLSAIR--HRFP-TTKDLFEAIVQC 533
D +L++ LQWIAA+ G Y F + +E L ++ HR D+F A+VQ
Sbjct 1621 DAELRATLQWIAASELGIPTIY--FKKSQENRIEKFLDVVQLVHRKSWKVGDIFHAVVQY 1678
Query 534 IQQREETPIGNWSLWQSLLNL 554
+ EE G SL+ LL L
Sbjct 1679 CKMHEEQKDGRLSLFDWLLEL 1699
> hsa:23424 TDRD7, KIAA1529, PCTAIRE2BP, RP11-508D10.1, TRAP;
tudor domain containing 7
Length=1098
Score = 33.1 bits (74), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 15/103 (14%)
Query 264 IISFYLHTPEATTFEAP------------EGEQVSRSEVDVSKIISQPLPLSGLTVDVHG 311
I+ + H P T E P + +Q+ RSE+D K+ PLP T + G
Sbjct 273 ILQQFEHWPHICTVEKPCSGGQDLLLYPAKRKQLLRSELDTEKVPLSPLPGPKQTPPLKG 332
Query 312 DITKSDGDLQVDFADMYVGGLS-MWAGHVAQ--EELIFALRPE 351
T GD + AD+ V S +WA + + EE+ PE
Sbjct 333 CPTVMAGDFKEKVADLLVKYTSGLWASALPKAFEEMYKVKFPE 375
> ath:AT3G07400 lipase class 3 family protein
Length=1003
Score = 33.1 bits (74), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 1/69 (1%)
Query 118 AQDEEFSRTTLPFLASVAKHISDLFPKGLKHMTRSHPSVHLRRIQVLCLMAASFFGIIPQ 177
A E S L SV I++L + H +S+ S +RI LC+ FFG+ Q
Sbjct 468 ASVESLSEGEYSKLTSVRSVITELRERLQSHSMKSYRS-RFQRIHDLCMDVDGFFGVDQQ 526
Query 178 KQRKLLRSW 186
KQ L+ W
Sbjct 527 KQFPHLQQW 535
> mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIAA1678;
SPHK1 interactor, AKAP domain containing
Length=1658
Score = 32.7 bits (73), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 44/83 (53%), Gaps = 9/83 (10%)
Query 477 DNQLKSLLQWIAATAAG-REMHYKAFGDATLAHMEGTLSAIRHRFPTTKDLFEAIVQ-C- 533
D +L++ LQWIAA+ G +++K ++ + + ++ + D+F A+VQ C
Sbjct 1579 DAELRATLQWIAASELGIPTIYFKKSQESRIEKFLDVVKLVQQKSWKVGDIFHAVVQYCK 1638
Query 534 --IQQREETPIGNWSLWQSLLNL 554
+Q+E TP SL+ LL L
Sbjct 1639 LHAEQKERTP----SLFDWLLEL 1657
Lambda K H
0.321 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 27696100120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40