bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0128_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
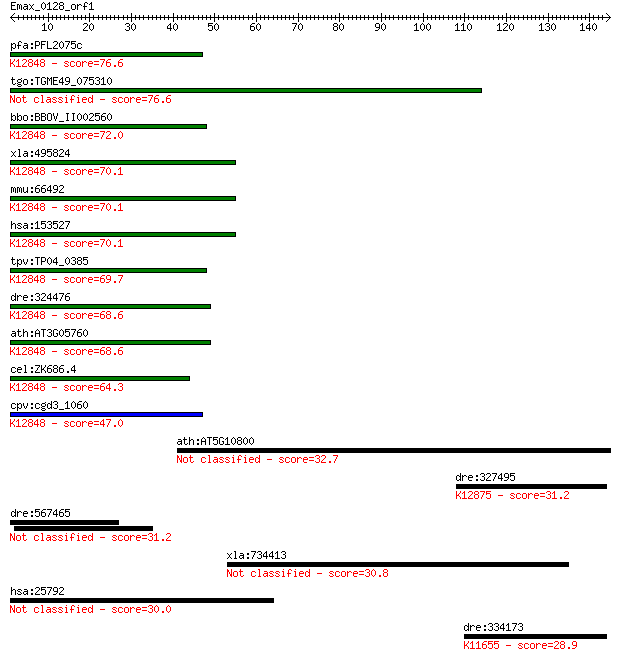
pfa:PFL2075c conserved Plasmodium protein; K12848 U4/U6.U5 tri... 76.6 3e-14
tgo:TGME49_075310 hypothetical protein 76.6 3e-14
bbo:BBOV_II002560 18.m06206; hypothetical protein; K12848 U4/U... 72.0 6e-13
xla:495824 zmat2, snu23; zinc finger, matrin-type 2; K12848 U4... 70.1 2e-12
mmu:66492 Zmat2, 2610510D14Rik, 2900082I05Rik; zinc finger, ma... 70.1 2e-12
hsa:153527 ZMAT2, FLJ31121, Snu23, hSNU23; zinc finger, matrin... 70.1 2e-12
tpv:TP04_0385 hypothetical protein; K12848 U4/U6.U5 tri-snRNP ... 69.7 3e-12
dre:324476 zmat2, fc30d10, flj31121, wu:fc30d10; zinc finger, ... 68.6 6e-12
ath:AT3G05760 nucleic acid binding / zinc ion binding; K12848 ... 68.6 6e-12
cel:ZK686.4 hypothetical protein; K12848 U4/U6.U5 tri-snRNP co... 64.3 1e-10
cpv:cgd3_1060 U1 like C2H2 zinc finger ; K12848 U4/U6.U5 tri-s... 47.0 2e-05
ath:AT5G10800 RNA recognition motif (RRM)-containing protein 32.7 0.44
dre:327495 acin1b, acinus, acinusb, fi14c05, wu:fe06a10, wu:fi... 31.2 1.2
dre:567465 zfr2, MGC174987, si:ch211-63j24.3, wu:fb99a09, zfr;... 31.2 1.3
xla:734413 larp1, MGC98945; La ribonucleoprotein domain family... 30.8 1.7
hsa:25792 CIZ1, LSFR1, NP94, ZNF356; CDKN1A interacting zinc f... 30.0 2.6
dre:334173 baz1a, fi34e04, wu:fi34e04; bromodomain adjacent to... 28.9 6.5
> pfa:PFL2075c conserved Plasmodium protein; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=242
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 29/46 (63%), Positives = 38/46 (82%), Gaps = 0/46 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAAL 46
+C IC+C+ KDSQ YLDHING+NHNR+LG SM+V++V+L VK L
Sbjct 113 YCKICDCVLKDSQTYLDHINGKNHNRMLGYSMKVKKVTLDDVKKRL 158
> tgo:TGME49_075310 hypothetical protein
Length=551
Score = 76.6 bits (187), Expect = 3e-14, Method: Composition-based stats.
Identities = 48/117 (41%), Positives = 67/117 (57%), Gaps = 4/117 (3%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQ--AERLALEAGGE 58
WCD+CECL KDS AY+DHINGRNHNR+LGM+MRVER + + V + L+ + LE GG+
Sbjct 364 WCDVCECLIKDSAAYMDHINGRNHNRMLGMTMRVERAAPSSVISRLREMQRQSQLELGGD 423
Query 59 QLNPKITVDAPLKQHQP--GGGGSQGAAGGIGSLPTGGAIEEGAPEAEDEFERAKRR 113
++ +K Q G +Q + + + G G E D+ +R KRR
Sbjct 424 DEGEAGSLSGDVKSQQSFDAPGAAQRGSVTRAAAASAGCAPGGIEENYDDIDRIKRR 480
> bbo:BBOV_II002560 18.m06206; hypothetical protein; K12848 U4/U6.U5
tri-snRNP component SNU23
Length=232
Score = 72.0 bits (175), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 37/47 (78%), Gaps = 0/47 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQ 47
+C C+C+ KDSQAY+DH+NGR HNR+LGM+MRVE+V V AL+
Sbjct 99 YCKTCDCILKDSQAYMDHLNGRKHNRMLGMTMRVEKVDHKAVAEALR 145
> xla:495824 zmat2, snu23; zinc finger, matrin-type 2; K12848
U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQAERLALE 54
+C++C+C+ KDS +LDHING+ H R LGMSMRVER +L +VK + + +E
Sbjct 81 YCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQVKKRFEVNKKKME 134
> mmu:66492 Zmat2, 2610510D14Rik, 2900082I05Rik; zinc finger,
matrin type 2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQAERLALE 54
+C++C+C+ KDS +LDHING+ H R LGMSMRVER +L +VK + + +E
Sbjct 81 YCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQVKKRFEVNKKKME 134
> hsa:153527 ZMAT2, FLJ31121, Snu23, hSNU23; zinc finger, matrin-type
2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQAERLALE 54
+C++C+C+ KDS +LDHING+ H R LGMSMRVER +L +VK + + +E
Sbjct 81 YCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQVKKRFEVNKKKME 134
> tpv:TP04_0385 hypothetical protein; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=259
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 36/47 (76%), Gaps = 0/47 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQ 47
+C C CL KDS++YLDH+NG+ HNRLLGM+MRVE+VS V L+
Sbjct 97 FCKTCNCLLKDSKSYLDHLNGKKHNRLLGMTMRVEKVSAKAVADKLR 143
> dre:324476 zmat2, fc30d10, flj31121, wu:fc30d10; zinc finger,
matrin type 2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=198
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQA 48
+C++C+C+ KDS +LDHING+ H R LGMSMRVER SL +VK +
Sbjct 80 YCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSSLDQVKKRFEV 127
> ath:AT3G05760 nucleic acid binding / zinc ion binding; K12848
U4/U6.U5 tri-snRNP component SNU23
Length=202
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQA 48
+C +C+C+ KDS YLDHING+ H R LGMSMRVER SL +V+ +
Sbjct 84 FCRVCDCVVKDSANYLDHINGKKHQRALGMSMRVERSSLEQVQERFEV 131
> cel:ZK686.4 hypothetical protein; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=217
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 22/43 (51%), Positives = 35/43 (81%), Gaps = 0/43 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVK 43
+CD+C+C+ KDS +LDHING+NH R +GMSM+ ++ ++A V+
Sbjct 91 YCDVCDCVVKDSINFLDHINGKNHQRNIGMSMKTKKSTVADVR 133
> cpv:cgd3_1060 U1 like C2H2 zinc finger ; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=198
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAAL 46
WC++C DS +++ H+N ++HN+ +G S+ VE+ SL VK L
Sbjct 84 WCEVCNLGFNDSHSWIRHLNSQSHNQKMGTSLYVEKKSLESVKRRL 129
> ath:AT5G10800 RNA recognition motif (RRM)-containing protein
Length=947
Score = 32.7 bits (73), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 56/116 (48%), Gaps = 12/116 (10%)
Query 41 RVKAALQAERLALEAGGEQLNPKITVDAPLKQHQPGGGGSQGAAGGIGS-LPTGGAIEEG 99
R K + RL LE + K+ D+ ++ + G S+ + + S LP A +
Sbjct 45 RFKGIMLQLRLLLEVTINPNDNKLKPDSQGEKSRDGDSISKKGSRYVPSFLPPPLASKGK 104
Query 100 APEAEDEFERAK-----------RRLEELEREQELKKQRKKERKKAAAAGGGNTAS 144
PE + + ER+K +EEL+REQE++++R ++R+ + NT+S
Sbjct 105 GPENKRDEERSKEMEKGKTRNIDHFVEELKREQEIRERRNQDRENSRDHNSDNTSS 160
> dre:327495 acin1b, acinus, acinusb, fi14c05, wu:fe06a10, wu:fi14c05;
apoptotic chromatin condensation inducer 1b; K12875
apoptotic chromatin condensation inducer in the nucleus
Length=1259
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 108 ERAKRRLEELEREQELKKQRKKERKKAAAAGGGNTA 143
ER KRR E E E + +K+R++ER K GGNTA
Sbjct 1159 EREKRRKEIFEEEDKKRKEREQERAKGRDRVGGNTA 1194
> dre:567465 zfr2, MGC174987, si:ch211-63j24.3, wu:fb99a09, zfr;
zinc finger RNA binding protein 2
Length=999
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 10/26 (38%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNR 26
+CDIC+ C Q Y +H+ G+ H +
Sbjct 314 YCDICKISCAGPQTYREHLEGQKHKK 339
Score = 28.9 bits (63), Expect = 6.1, Method: Composition-based stats.
Identities = 9/33 (27%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 2 CDICECLCKDSQAYLDHINGRNHNRLLGMSMRV 34
C++C+ C + AY HI G H +++ + ++
Sbjct 363 CELCDVSCTGADAYAAHIRGSKHQKVVKLHTKL 395
> xla:734413 larp1, MGC98945; La ribonucleoprotein domain family,
member 1
Length=1021
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 20/89 (22%), Positives = 42/89 (47%), Gaps = 7/89 (7%)
Query 53 LEAGGEQLNPKITVDAPLKQH-------QPGGGGSQGAAGGIGSLPTGGAIEEGAPEAED 105
++AGG +N ++ VD P+ +P G G G + PT G I + +
Sbjct 69 MKAGGSTVNGQLPVDPPVPAKVVKAGNPRPRRGSKVGDFGDATNWPTPGEIAHKSVQPSS 128
Query 106 EFERAKRRLEELEREQELKKQRKKERKKA 134
+ + +++ L + E ++E ++ +R K+
Sbjct 129 KLQPSRKSLGKKEMKEESADGKENQRVKS 157
> hsa:25792 CIZ1, LSFR1, NP94, ZNF356; CDKN1A interacting zinc
finger protein 1
Length=842
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 31/63 (49%), Gaps = 3/63 (4%)
Query 1 WCDICECLCKDSQAYLDHINGRNHNRLLGMSMRVERVSLARVKAALQAERLALEAGGEQL 60
+C IC+ C Q + DH++ H + LG ++ +S A + + L R LE E+
Sbjct 538 FCYICKASCSSQQEFQDHMSEPQHQQRLG---EIQHMSQACLLSLLPVPRDVLETEDEEP 594
Query 61 NPK 63
P+
Sbjct 595 PPR 597
> dre:334173 baz1a, fi34e04, wu:fi34e04; bromodomain adjacent
to zinc finger domain, 1A; K11655 bromodomain adjacent to zinc
finger domain protein 1A
Length=711
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 110 AKRRLEELEREQELKKQRKKERKKAAAAGGGNTA 143
++R EE REQELK + K ER K A GG A
Sbjct 647 VRKRKEEKLREQELKLKEKHERLKEEAKNGGQLA 680
Lambda K H
0.312 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40