bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0116_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
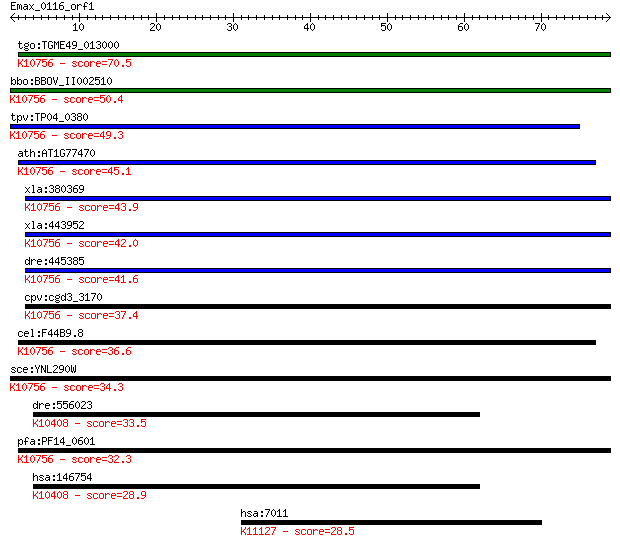
tgo:TGME49_013000 activator 1 36 kDa, putative (EC:2.7.7.7); K... 70.5 1e-12
bbo:BBOV_II002510 18.m06203; replication factor C3 protein; K1... 50.4 1e-06
tpv:TP04_0380 replication factor C subunit 3; K10756 replicati... 49.3 3e-06
ath:AT1G77470 replication factor C 36 kDA, putative; K10756 re... 45.1 5e-05
xla:380369 rfc5, MGC53482; replication factor C (activator 1) ... 43.9 1e-04
xla:443952 MGC80325 protein; K10756 replication factor C subun... 42.0 5e-04
dre:445385 rfc5, zgc:110313; replication factor C (activator 1... 41.6 6e-04
cpv:cgd3_3170 replication factor RFC3 AAA+ ATpase ; K10756 rep... 37.4 0.011
cel:F44B9.8 ARPA; hypothetical protein; K10756 replication fac... 36.6 0.020
sce:YNL290W RFC3; Rfc3p; K10756 replication factor C subunit 3/5 34.3
dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein... 33.5 0.16
pfa:PF14_0601 replication factor C3; K10756 replication factor... 32.3 0.36
hsa:146754 DNAH2, DNAHC2, DNHD3, FLJ46675, KIAA1503; dynein, a... 28.9 4.7
hsa:7011 TEP1, TLP1, TP1, TROVE1, VAULT2, p240; telomerase-ass... 28.5 5.2
> tgo:TGME49_013000 activator 1 36 kDa, putative (EC:2.7.7.7);
K10756 replication factor C subunit 3/5
Length=398
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 35/77 (45%), Positives = 49/77 (63%), Gaps = 0/77 (0%)
Query 2 PSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVT 61
PSEV ++ + LL D C E LVT +GY++RDWV A H ++ + P V +T V+
Sbjct 293 PSEVTTMFERLLVADFFACCKELDELVTAKGYAMRDWVIAFHERILLVDWPANVLITFVS 352
Query 62 RLADIEERLALGSGDYV 78
RLAD+EERLA G+ + V
Sbjct 353 RLADLEERLATGASEAV 369
> bbo:BBOV_II002510 18.m06203; replication factor C3 protein;
K10756 replication factor C subunit 3/5
Length=348
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 1 QPSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLV 60
P+E+ ++Q L+++ + C L +GYSV D V AL+ + + P V + L+
Sbjct 245 NPTEISKLLQRLMQESFKDCVDYVVTLNQVQGYSVEDLVTALYRSILRIDWPNVVIVQLL 304
Query 61 TRLADIEERLALGSGDYV 78
RL DIE+RL+ G+ Y+
Sbjct 305 IRLGDIEQRLSAGASPYI 322
> tpv:TP04_0380 replication factor C subunit 3; K10756 replication
factor C subunit 3/5
Length=347
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 45/75 (60%), Gaps = 2/75 (2%)
Query 1 QPSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLP-VPVALTL 59
Q SE+ +++ L++ + C E L ++GYS+ D V L+ + + P VP+ + L
Sbjct 244 QSSEIDHLLKSLMQNSFKECIYELSVLHHKKGYSLEDIVRLLYKSIVKIDWPNVPI-VQL 302
Query 60 VTRLADIEERLALGS 74
+ RLAD+EERLA G+
Sbjct 303 LIRLADVEERLAAGA 317
> ath:AT1G77470 replication factor C 36 kDA, putative; K10756
replication factor C subunit 3/5
Length=369
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 2 PSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVT 61
P +++ I LL K C + + TR+G ++ D V + + +K+P V + L+
Sbjct 272 PKDIEQISHWLLNKPFDECYKDVSEIKTRKGLAIVDIVKEITLFIFKIKMPSAVRVQLIN 331
Query 62 RLADIEERLALGSGD 76
LADIE RL+ G D
Sbjct 332 DLADIEYRLSFGCND 346
> xla:380369 rfc5, MGC53482; replication factor C (activator 1)
5, 36.5kDa; K10756 replication factor C subunit 3/5
Length=335
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 SEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVTR 62
S++ +I+ +L KD + L T +G ++ D + +H + + P V + L+ +
Sbjct 240 SDIANILDWMLNKDFTSAYKNIMELKTLKGLALHDILTEVHLYVHRVNFPASVRMHLLVK 299
Query 63 LADIEERLALGSGDYV 78
+ADIE RLA G+ + +
Sbjct 300 MADIEYRLASGTSEKI 315
> xla:443952 MGC80325 protein; K10756 replication factor C subunit
3/5
Length=335
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 SEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVTR 62
S++ +I+ +L KD + L T +G ++ D + +H + + P V + L+ +
Sbjct 240 SDIANILDWMLNKDFTSAYKNIMELKTLKGLALHDILTEIHLYVHRVDFPALVRIHLLVK 299
Query 63 LADIEERLALGSGDYV 78
+ADIE RLA G+ + +
Sbjct 300 MADIEYRLASGTSEKI 315
> dre:445385 rfc5, zgc:110313; replication factor C (activator
1) 5; K10756 replication factor C subunit 3/5
Length=334
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 SEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVTR 62
S++ +I+ L KD T + L T +G ++ D + +H + + P + + L+ +
Sbjct 239 SDIANILDWALNKDFTTAYNQILELKTLKGLALHDILTEVHLLIHRVDFPPSIRMGLLIK 298
Query 63 LADIEERLALGSGDYV 78
LADIE RLA G+ + +
Sbjct 299 LADIEYRLASGTSEKI 314
> cpv:cgd3_3170 replication factor RFC3 AAA+ ATpase ; K10756 replication
factor C subunit 3/5
Length=383
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 35/76 (46%), Gaps = 0/76 (0%)
Query 3 SEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVTR 62
SE+ I +L + + GYS +D+V L+ + P V L+ R
Sbjct 288 SELDYIFGILSRESFSSGFSALQNSQNENGYSTQDFVNGLYSKSMEANWPDEVVPLLMRR 347
Query 63 LADIEERLALGSGDYV 78
LADIE RL+ G+ + +
Sbjct 348 LADIEYRLSRGASESI 363
> cel:F44B9.8 ARPA; hypothetical protein; K10756 replication factor
C subunit 3/5
Length=368
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 37/75 (49%), Gaps = 0/75 (0%)
Query 2 PSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVT 61
P E+K +++ LL + C + GY+++D + LH + + +P ++T
Sbjct 262 PKEMKEVVKTLLNDPSKKCMNTIQTKLFENGYALQDVITHLHDFVFTLDIPDEAMSAIIT 321
Query 62 RLADIEERLALGSGD 76
L ++EE L+ G +
Sbjct 322 GLGEVEENLSTGCSN 336
> sce:YNL290W RFC3; Rfc3p; K10756 replication factor C subunit
3/5
Length=340
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 19/79 (24%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Query 1 QPSEVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLP-VPVALTL 59
+PS++K++++ +LE D T + + + +G ++ D + + +++ +L + L
Sbjct 240 RPSDLKAVLKSILEDDWGTAHYTLNKVRSAKGLALIDLIEGIVKILEDYELQNEETRVHL 299
Query 60 VTRLADIEERLALGSGDYV 78
+T+LADIE ++ G D +
Sbjct 300 LTKLADIEYSISKGGNDQI 318
> dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein
heavy chain, axonemal
Length=4424
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 4 EVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVT 61
+V+ M+ L+ LR CS+ + +RG VRDW G + ++ V L+T
Sbjct 1607 DVERTMRWTLKDSLRNCSMALKKMPGKRGKWVRDWPGQMLITASQIQWTTDVTRALMT 1664
> pfa:PF14_0601 replication factor C3; K10756 replication factor
C subunit 3/5
Length=344
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Query 2 PSEVKSIMQVLLEKDLRTCSLEF-HGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLV 60
PSE K I++ + ++ S EF L +GYS +D + L+ + P L+
Sbjct 242 PSETKKILEYFTKGSIKE-SYEFVSNLQYDKGYSTKDIMMCLYESVLTYDFPDSAFCLLL 300
Query 61 TRLADIEERLALGSGDYV 78
+IEER + G+ + +
Sbjct 301 KNFGEIEERCSSGASEQI 318
> hsa:146754 DNAH2, DNAHC2, DNHD3, FLJ46675, KIAA1503; dynein,
axonemal, heavy chain 2; K10408 dynein heavy chain, axonemal
Length=4427
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 4 EVKSIMQVLLEKDLRTCSLEFHGLVTRRGYSVRDWVGALHCQMQNMKLPVPVALTLVT 61
+V+ M+V L LR C L + +R V++W G + ++ V L+T
Sbjct 1612 DVEQTMRVTLRDLLRNCHLALRKFLNKRDKWVKEWAGQVVITASQIQWTADVTKCLLT 1669
> hsa:7011 TEP1, TLP1, TP1, TROVE1, VAULT2, p240; telomerase-associated
protein 1; K11127 telomerase protein component 1
Length=2627
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 31 RGYSVRDWVGALHCQMQNMKLPVPVALTLVTRLADIEER 69
R + D + AL Q++N LP P +TL+ R+ E+
Sbjct 565 RFLNAHDAIDALEAQLRNQALPFPSNITLMRRILTRNEK 603
Lambda K H
0.323 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40