bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0077_orf1
Length=189
Score E
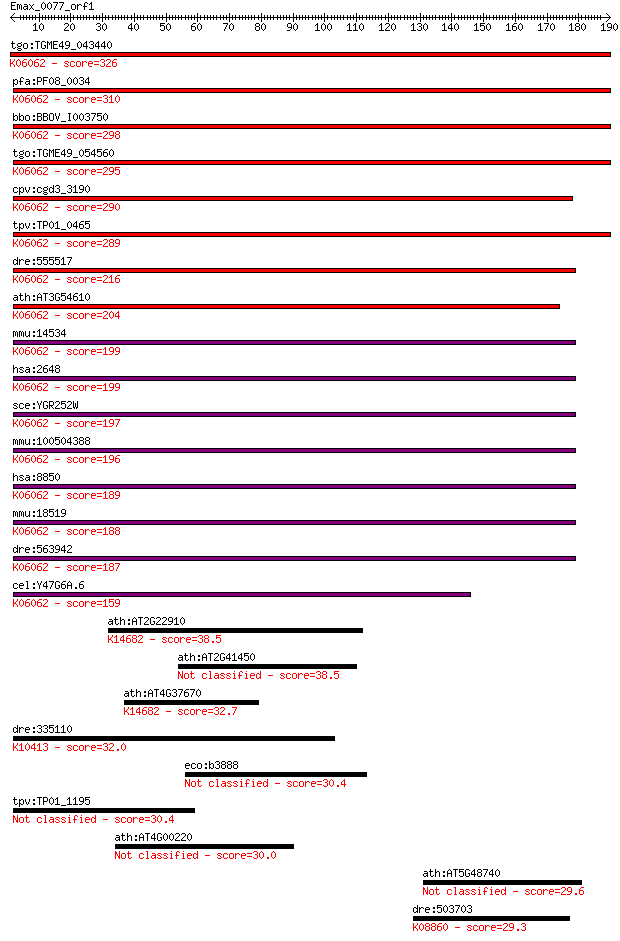
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_043440 histone acetyltransferase GCN5, putative (EC... 326 3e-89
pfa:PF08_0034 gcn5; histone acetyltransferase GCN5, putative; ... 310 1e-84
bbo:BBOV_I003750 19.m02034; histone acetyltransferase; K06062 ... 298 7e-81
tgo:TGME49_054560 histone acetyltransferase, putative ; K06062... 295 9e-80
cpv:cgd3_3190 GCN5 like acetylase + bromodomain ; K06062 histo... 290 2e-78
tpv:TP01_0465 histone acetyltransferase Gcn5; K06062 histone a... 289 3e-78
dre:555517 kat2a, gb:dq017634, im:7156024; K(lysine) acetyltra... 216 4e-56
ath:AT3G54610 HAG1; GCN5; DNA binding / H3 histone acetyltrans... 204 1e-52
mmu:14534 Kat2a, 1110051E14Rik, AW212720, Gcn5, Gcn5l2, mmGCN5... 199 7e-51
hsa:2648 KAT2A, GCN5, GCN5L2, MGC102791, PCAF-b, hGCN5; K(lysi... 199 7e-51
sce:YGR252W GCN5, AAS104, ADA4, SWI9; Gcn5p (EC:2.3.1.48); K06... 197 1e-50
mmu:100504388 Kat2b-ps, EG330129, Gm5109, Pcaf-ps; K(lysine) a... 196 4e-50
hsa:8850 KAT2B, CAF, P, P/CAF, PCAF; K(lysine) acetyltransfera... 189 3e-48
mmu:18519 Kat2b, A930006P13Rik, AI461839, AW536563, Pcaf, p/CA... 188 8e-48
dre:563942 kat2b, MGC161980, pcaf, si:ch211-1j13.2, zgc:161980... 187 2e-47
cel:Y47G6A.6 pcaf-1; P300/CBP Associated Factor homolog family... 159 4e-39
ath:AT2G22910 GCN5-related N-acetyltransferase (GNAT) family p... 38.5 0.013
ath:AT2G41450 N-acetyltransferase 38.5 0.015
ath:AT4G37670 GCN5-related N-acetyltransferase (GNAT) family p... 32.7 0.83
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 32.0 1.3
eco:b3888 yiiD, ECK3881, JW3859; predicted acetyltransferase 30.4 3.3
tpv:TP01_1195 cop-coated vesicle membrane protein p24 precursor 30.4 3.8
ath:AT4G00220 JLO; JLO (JAGGED LATERAL ORGANS) 30.0 4.7
ath:AT5G48740 leucine-rich repeat family protein / protein kin... 29.6 7.0
dre:503703 pkz, MGC136657, MGC158143, zgc:136657; protein kina... 29.3 8.4
> tgo:TGME49_043440 histone acetyltransferase GCN5, putative (EC:4.1.1.70);
K06062 histone acetyltransferase [EC:2.3.1.48]
Length=1032
Score = 326 bits (835), Expect = 3e-89, Method: Compositional matrix adjust.
Identities = 147/189 (77%), Positives = 170/189 (89%), Gaps = 0/189 (0%)
Query 1 ALVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAF 60
ALV KNIFSRQLPKMPREYIVRLVFDRNHYTFCL K+ IIGGCCFRPYF+QKFAE+AF
Sbjct 647 ALVTVKNIFSRQLPKMPREYIVRLVFDRNHYTFCLNKEDTIIGGCCFRPYFQQKFAEIAF 706
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERW 120
LAVTS EQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMP+ERW
Sbjct 707 LAVTSTEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPRERW 766
Query 121 FGFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQEDP 180
+G+IKDYEGGTLMEC I+P+++YLRL +L+DQQ+ ++RA + LKP V+PGL W+++P
Sbjct 767 YGYIKDYEGGTLMECHINPRINYLRLSEMLHDQQQVIKRATVSLKPLAVYPGLDFWKKNP 826
Query 181 TRVLHPSEV 189
+ L PS++
Sbjct 827 GQTLSPSQI 835
> pfa:PF08_0034 gcn5; histone acetyltransferase GCN5, putative;
K06062 histone acetyltransferase [EC:2.3.1.48]
Length=1465
Score = 310 bits (795), Expect = 1e-84, Method: Composition-based stats.
Identities = 136/188 (72%), Positives = 162/188 (86%), Gaps = 0/188 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
L+ KNIFSRQLPKMPREYIVRLVFDRNHYTFCL+K+ +IGG CFRPYF+QKFAE+AFL
Sbjct 1148 LITLKNIFSRQLPKMPREYIVRLVFDRNHYTFCLLKKNTVIGGVCFRPYFEQKFAEIAFL 1207
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGTRLMNHLKEHVKK GIEYFLTYADNFA+GYFRKQGF+QKISMPKERWF
Sbjct 1208 AVTSTEQVKGYGTRLMNHLKEHVKKFGIEYFLTYADNFAIGYFRKQGFSQKISMPKERWF 1267
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQEDPT 181
G+IKDY+GGTLMEC I P ++YLRL +L +Q++AV++AI +KP ++ G+ + ++
Sbjct 1268 GYIKDYDGGTLMECYIFPNINYLRLSEMLYEQKKAVKKAIHFIKPQVIYKGINYFADNKG 1327
Query 182 RVLHPSEV 189
LHPS +
Sbjct 1328 AALHPSTI 1335
> bbo:BBOV_I003750 19.m02034; histone acetyltransferase; K06062
histone acetyltransferase [EC:2.3.1.48]
Length=646
Score = 298 bits (763), Expect = 7e-81, Method: Compositional matrix adjust.
Identities = 134/188 (71%), Positives = 162/188 (86%), Gaps = 0/188 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV KNIFSRQLPKMPREYIVRLVFDRNHYTFCL+K+ ++IGG CFRPYF+Q+FAE+AFL
Sbjct 323 LVTVKNIFSRQLPKMPREYIVRLVFDRNHYTFCLLKKGEVIGGICFRPYFEQRFAEIAFL 382
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AV S EQVKGYGTR+MNHLKEHVKKS IEYFLTYADNFA+GYFRKQGF+QKISMPKERWF
Sbjct 383 AVKSTEQVKGYGTRIMNHLKEHVKKSNIEYFLTYADNFAIGYFRKQGFSQKISMPKERWF 442
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQEDPT 181
G+IKDY+GGTLMEC ISP ++YLRL +L Q+ + + I +KP +V+ GLT ++E+P
Sbjct 443 GYIKDYDGGTLMECYISPNINYLRLSDMLGKQKAIISQCIEAIKPLKVYDGLTFFKENPG 502
Query 182 RVLHPSEV 189
++P ++
Sbjct 503 ITINPRDI 510
> tgo:TGME49_054560 histone acetyltransferase, putative ; K06062
histone acetyltransferase [EC:2.3.1.48]
Length=447
Score = 295 bits (754), Expect = 9e-80, Method: Compositional matrix adjust.
Identities = 134/188 (71%), Positives = 161/188 (85%), Gaps = 0/188 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV KNIFSRQLPKMPREYIVRLVFDR H+TFCL KQ ++IGG CFRPYF++KFAE+AFL
Sbjct 121 LVTVKNIFSRQLPKMPREYIVRLVFDRAHFTFCLCKQGRVIGGVCFRPYFREKFAEIAFL 180
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGF+ KI+MP++RW
Sbjct 181 AVTSTEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFSSKITMPRDRWL 240
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQEDPT 181
G+IKDY+GGTLMECR+S +++YL+L +L Q+ AV+R I + P V P L+ W+E+P
Sbjct 241 GYIKDYDGGTLMECRLSTRINYLKLSQLLALQKLAVKRRIEQSAPSVVCPSLSFWKENPG 300
Query 182 RVLHPSEV 189
++L PS +
Sbjct 301 QLLMPSAI 308
> cpv:cgd3_3190 GCN5 like acetylase + bromodomain ; K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=655
Score = 290 bits (742), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 125/176 (71%), Positives = 158/176 (89%), Gaps = 0/176 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
L+A KN+F+RQLPKMPREYIVRL+FDRNHY+FCL+K+ ++IGG CFRPYF+Q+FAE+AFL
Sbjct 281 LIALKNVFARQLPKMPREYIVRLIFDRNHYSFCLLKKGKVIGGVCFRPYFEQRFAEIAFL 340
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGTRLMNHLK+HVKKS IEYFLTYADNFA GYFRKQGF +++SMPKERWF
Sbjct 341 AVTSTEQVKGYGTRLMNHLKQHVKKSKIEYFLTYADNFATGYFRKQGFRKEVSMPKERWF 400
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQ 177
G+IKDY+GGTLMEC I+P+++YLRL + ++Q+ A+ RAI ++P +V+PG+ W+
Sbjct 401 GYIKDYDGGTLMECYINPEINYLRLSDLFHEQKSALLRAINTIRPLKVYPGINLWE 456
> tpv:TP01_0465 histone acetyltransferase Gcn5; K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=631
Score = 289 bits (740), Expect = 3e-78, Method: Compositional matrix adjust.
Identities = 127/188 (67%), Positives = 160/188 (85%), Gaps = 0/188 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
L+ KNIFSRQLPKMPREYIVRLVFDRNHYTFCL+K+ ++IGG CFRPYF+Q+FAE+AFL
Sbjct 311 LITVKNIFSRQLPKMPREYIVRLVFDRNHYTFCLLKKGEVIGGICFRPYFEQRFAEIAFL 370
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AV S EQ+KGYGTR+MNHLKEHVKKS IEYFLTYADNFA+GYF+KQGF+ KI+MP+ERWF
Sbjct 371 AVKSTEQIKGYGTRIMNHLKEHVKKSNIEYFLTYADNFAIGYFKKQGFSLKITMPRERWF 430
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTCWQEDPT 181
G+IKDY+GGTLMEC ISP ++YLRL +L+ Q+ V + I +KP +V+ GL + ++ T
Sbjct 431 GYIKDYDGGTLMECYISPNINYLRLSDMLSQQKAIVVKCIEAIKPLKVYSGLNVFNKNTT 490
Query 182 RVLHPSEV 189
++P ++
Sbjct 491 VTINPCDI 498
> dre:555517 kat2a, gb:dq017634, im:7156024; K(lysine) acetyltransferase
2A; K06062 histone acetyltransferase [EC:2.3.1.48]
Length=795
Score = 216 bits (550), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 101/179 (56%), Positives = 132/179 (73%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 478 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLALIKDGRVIGGICFRMFPTQGFTEIVFC 537
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K GI YFLTYAD +A+GYF+KQGF++ I +PK R+
Sbjct 538 AVTSNEQVKGYGTHLMNHLKEYHIKHGILYFLTYADEYAIGYFKKQGFSKDIKVPKSRYL 597
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLMEC ++P++ Y L I+ Q+E +++ I R + +V+PGLTC++E
Sbjct 598 GYIKDYEGATLMECELNPRIPYTELSHIIKRQKEIIKKLIERKQNQIRKVYPGLTCFKE 656
> ath:AT3G54610 HAG1; GCN5; DNA binding / H3 histone acetyltransferase/
histone acetyltransferase; K06062 histone acetyltransferase
[EC:2.3.1.48]
Length=568
Score = 204 bits (520), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 93/174 (53%), Positives = 126/174 (72%), Gaps = 2/174 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
L+ KNIF+RQLP MP+EYIVRL+ DR H + +++ ++GG +RPY QKF E+AF
Sbjct 234 LIGLKNIFARQLPNMPKEYIVRLLMDRKHKSVMVLRGNLVVGGITYRPYHSQKFGEIAFC 293
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKK-SGIEYFLTYADNFAVGYFRKQGFTQKISMPKERW 120
A+T+ EQVKGYGTRLMNHLK+H + G+ +FLTYADN AVGYF KQGFT++I + K+ W
Sbjct 294 AITADEQVKGYGTRLMNHLKQHARDVDGLTHFLTYADNNAVGYFVKQGFTKEIYLEKDVW 353
Query 121 FGFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKP-PRVFPGL 173
GFIKDY+GG LMEC+I PK+ Y L +++ Q++A+ I L V+P +
Sbjct 354 HGFIKDYDGGLLMECKIDPKLPYTDLSSMIRQQRKAIDERIRELSNCQNVYPKI 407
> mmu:14534 Kat2a, 1110051E14Rik, AW212720, Gcn5, Gcn5l2, mmGCN5;
K(lysine) acetyltransferase 2A (EC:2.3.1.48); K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=829
Score = 199 bits (505), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 99/179 (55%), Positives = 131/179 (73%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 512 LVGLQNVFSHQLPRMPKEYIARLVFDPKHKTLALIKDGRVIGGICFRMFPTQGFTEIVFC 571
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I YFLTYAD +A+GYF+KQGF++ I +PK R+
Sbjct 572 AVTSNEQVKGYGTHLMNHLKEYHIKHSILYFLTYADEYAIGYFKKQGFSKDIKVPKSRYL 631
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLMEC ++P++ Y L I+ Q+E +++ I R + +V+PGL+C++E
Sbjct 632 GYIKDYEGATLMECELNPRIPYTELSHIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKE 690
> hsa:2648 KAT2A, GCN5, GCN5L2, MGC102791, PCAF-b, hGCN5; K(lysine)
acetyltransferase 2A (EC:2.3.1.48); K06062 histone acetyltransferase
[EC:2.3.1.48]
Length=837
Score = 199 bits (505), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 99/179 (55%), Positives = 131/179 (73%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 520 LVGLQNVFSHQLPRMPKEYIARLVFDPKHKTLALIKDGRVIGGICFRMFPTQGFTEIVFC 579
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I YFLTYAD +A+GYF+KQGF++ I +PK R+
Sbjct 580 AVTSNEQVKGYGTHLMNHLKEYHIKHNILYFLTYADEYAIGYFKKQGFSKDIKVPKSRYL 639
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLMEC ++P++ Y L I+ Q+E +++ I R + +V+PGL+C++E
Sbjct 640 GYIKDYEGATLMECELNPRIPYTELSHIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKE 698
> sce:YGR252W GCN5, AAS104, ADA4, SWI9; Gcn5p (EC:2.3.1.48); K06062
histone acetyltransferase [EC:2.3.1.48]
Length=439
Score = 197 bits (502), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 91/180 (50%), Positives = 133/180 (73%), Gaps = 3/180 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQ-QIIGGCCFRPYFKQKFAEVAF 60
L KNIF +QLPKMP+EYI RLV+DR+H + +I++ ++GG +RP+ K++FAE+ F
Sbjct 117 LTGLKNIFQKQLPKMPKEYIARLVYDRSHLSMAVIRKPLTVVGGITYRPFDKREFAEIVF 176
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKK-SGIEYFLTYADNFAVGYFRKQGFTQKISMPKER 119
A++S EQV+GYG LMNHLK++V+ S I+YFLTYADN+A+GYF+KQGFT++I++ K
Sbjct 177 CAISSTEQVRGYGAHLMNHLKDYVRNTSNIKYFLTYADNYAIGYFKKQGFTKEITLDKSI 236
Query 120 WFGFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVF-PGLTCWQE 178
W G+IKDYEGGTLM+C + P++ YL IL Q+ A++R I + + PGL +++
Sbjct 237 WMGYIKDYEGGTLMQCSMLPRIRYLDAGKILLLQEAALRRKIRTISKSHIVRPGLEQFKD 296
> mmu:100504388 Kat2b-ps, EG330129, Gm5109, Pcaf-ps; K(lysine)
acetyltransferase 2B, pseudogene; K06062 histone acetyltransferase
[EC:2.3.1.48]
Length=813
Score = 196 bits (498), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 92/179 (51%), Positives = 126/179 (70%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CF+ + Q F E+ F
Sbjct 496 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLALIKDGRVIGGICFQMFPSQGFTEIVFC 555
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I FLTYAD+ A+GYF+KQGF+++I +PK ++
Sbjct 556 AVTSEEQVKGYGTHLMNHLKEYHVKHEILNFLTYADDHAIGYFKKQGFSKEIKIPKTKYA 615
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLM C ++P + Y I+ Q+E ++ R + +V+PGL+C+++
Sbjct 616 GYIKDYEGATLMGCELNPHIPYTEFSVIIKKQKEITKKLTERKQAQIGKVYPGLSCFKD 674
> hsa:8850 KAT2B, CAF, P, P/CAF, PCAF; K(lysine) acetyltransferase
2B (EC:2.3.1.48); K06062 histone acetyltransferase [EC:2.3.1.48]
Length=832
Score = 189 bits (481), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 92/180 (51%), Positives = 130/180 (72%), Gaps = 4/180 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 515 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLALIKDGRVIGGICFRMFPSQGFTEIVFC 574
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I FLTYAD +A+GYF+KQGF+++I +PK ++
Sbjct 575 AVTSNEQVKGYGTHLMNHLKEYHIKHDILNFLTYADEYAIGYFKKQGFSKEIKIPKTKYV 634
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP---RVFPGLTCWQE 178
G+IKDYEG TLM C ++P++ Y F+++ +Q+ + + ++ K +V+PGL+C+++
Sbjct 635 GYIKDYEGATLMGCELNPRIPYTE-FSVIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKD 693
> mmu:18519 Kat2b, A930006P13Rik, AI461839, AW536563, Pcaf, p/CAF;
K(lysine) acetyltransferase 2B (EC:2.3.1.48); K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=735
Score = 188 bits (478), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 92/180 (51%), Positives = 130/180 (72%), Gaps = 4/180 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 418 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLALIKDGRVIGGICFRMFPSQGFTEIVFC 477
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I FLTYAD +A+GYF+KQGF+++I +PK ++
Sbjct 478 AVTSNEQVKGYGTHLMNHLKEYHIKHEILNFLTYADEYAIGYFKKQGFSKEIKIPKTKYV 537
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP---RVFPGLTCWQE 178
G+IKDYEG TLM C ++P++ Y F+++ +Q+ + + ++ K +V+PGL+C+++
Sbjct 538 GYIKDYEGATLMGCELNPQIPYTE-FSVIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKD 596
> dre:563942 kat2b, MGC161980, pcaf, si:ch211-1j13.2, zgc:161980;
K(lysine) acetyltransferase 2B (EC:2.3.1.48); K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=796
Score = 187 bits (475), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 93/180 (51%), Positives = 128/180 (71%), Gaps = 4/180 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 481 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLSLIKDGRVIGGICFRMFPTQGFTEIVFC 540
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I FLTYAD +A+GYF+KQGF++ I +PK ++
Sbjct 541 AVTSNEQVKGYGTHLMNHLKEYHIKHEILNFLTYADEYAIGYFKKQGFSKDIKVPKSKYV 600
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP---RVFPGLTCWQE 178
G+IKDYEG TLM C ++P + Y F+++ +Q+ + + ++ K +V+PGL+C++E
Sbjct 601 GYIKDYEGATLMGCELNPCIPYTE-FSVIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKE 659
> cel:Y47G6A.6 pcaf-1; P300/CBP Associated Factor homolog family
member (pcaf-1); K06062 histone acetyltransferase [EC:2.3.1.48]
Length=767
Score = 159 bits (403), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 71/145 (48%), Positives = 102/145 (70%), Gaps = 1/145 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQ-IIGGCCFRPYFKQKFAEVAF 60
LV +N+F QLPKMP+EY+ RL+FD H ++K+ +IGG CFR + + F E+ F
Sbjct 436 LVELQNLFGAQLPKMPKEYVTRLIFDSRHQNMVILKRDMGVIGGICFRTFPSRGFVEIVF 495
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERW 120
A+T+ EQVKGYGT LMNH K+++ K+ I + LTYAD FA+GYF KQGF++K+ + +
Sbjct 496 CAITAMEQVKGYGTHLMNHCKDYMIKNKIYHMLTYADEFAIGYFTKQGFSEKLEINDTVY 555
Query 121 FGFIKDYEGGTLMECRISPKVDYLR 145
G+IK+YEG TLM C + P++ Y +
Sbjct 556 QGWIKEYEGATLMGCHLHPQISYTK 580
> ath:AT2G22910 GCN5-related N-acetyltransferase (GNAT) family
protein / amino acid kinase family protein (EC:2.7.2.8); K14682
amino-acid N-acetyltransferase [EC:2.3.1.1]
Length=609
Score = 38.5 bits (88), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 32 TFCLIKQQ-QIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYGTRLMNHLKEHVKKSGIE 90
+F +++++ II P+F++K EVA +AV S + +G G +L++++++ G+E
Sbjct 486 SFVVVEREGHIIACAALFPFFEEKCGEVAAIAVASDCRGQGQGDKLLDYIEKKASALGLE 545
Query 91 YFLTYADNFAVGYFRKQGFTQ 111
A +F ++GF +
Sbjct 546 MLFLLTTRTA-DWFVRRGFQE 565
> ath:AT2G41450 N-acetyltransferase
Length=549
Score = 38.5 bits (88), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 54 KFAEVAFLAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGF 109
++AE+ AVT Q KG+G + L + + GI +AD + G++ KQGF
Sbjct 84 QYAEIPLAAVTYTHQKKGFGKLVYEELMKRLHSVGIRTIYCWADKESEGFWLKQGF 139
> ath:AT4G37670 GCN5-related N-acetyltransferase (GNAT) family
protein / amino acid kinase family protein; K14682 amino-acid
N-acetyltransferase [EC:2.3.1.1]
Length=543
Score = 32.7 bits (73), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 37 KQQQIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYGTRLMN 78
++ QII P+FK K EVA +AV S + +G G +L+
Sbjct 501 REGQIIACAALFPFFKDKCGEVAAIAVASDCRGQGQGDKLLG 542
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 45/104 (43%), Gaps = 3/104 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVR-LVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAF 60
L++A N+ ++ K+ RE R D N L +Q+ +I C K ++
Sbjct 2096 LISAGNVKRERIQKIKREKCERGEDVDENEIAENLPEQEILIQSVCETMVPKLVAEDIPL 2155
Query 61 LAVTSAEQVKG--YGTRLMNHLKEHVKKSGIEYFLTYADNFAVG 102
L ++ G Y M L+E +KK E +LTY D VG
Sbjct 2156 LFSLLSDVFPGVQYMRGEMTALREELKKVCAEMYLTYGDGDDVG 2199
> eco:b3888 yiiD, ECK3881, JW3859; predicted acetyltransferase
Length=329
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 56 AEVAFLAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQK 112
A + F+AV Q KG GT + L+ ++ G++ A AV +F K GF +
Sbjct 85 ASIRFMAVHPDVQDKGLGTLMAMTLESVARQEGVKRVTCSAREDAVEFFAKLGFVNQ 141
> tpv:TP01_1195 cop-coated vesicle membrane protein p24 precursor
Length=211
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNH---YTFCLIKQQQIIGGCCFRPYFKQKFAEV 58
L + + I R REY++ + ++ + T+ +++ +IG CC++ Y F EV
Sbjct 148 LTSTREIMERMEAYSTREYLLSKIINKMNSRIITWSIVQMIVVIGLCCYQIYHISSFFEV 207
> ath:AT4G00220 JLO; JLO (JAGGED LATERAL ORGANS)
Length=228
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 34 CLIKQQQIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYG--TRLMNHLKEHVKKSGI 89
C +++ + GC F PYF + F AV +V G ++L++H+ EH + +
Sbjct 21 CKFLRRKCVAGCIFAPYFDSEQGAAHFAAV---HKVFGASNVSKLLHHVPEHKRPDAV 75
> ath:AT5G48740 leucine-rich repeat family protein / protein kinase
family protein
Length=895
Score = 29.6 bits (65), Expect = 7.0, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 131 TLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTC-WQEDP 180
TL + + +P+V L ++ IL EA + LK F G WQ+DP
Sbjct 320 TLRKIKFNPQVSALEVYEILQIPPEASSTTVSALKVIEQFTGQDLGWQDDP 370
> dre:503703 pkz, MGC136657, MGC158143, zgc:136657; protein kinase
containing Z-DNA binding domains; K08860 eukaryotic translation
initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=511
Score = 29.3 bits (64), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 27/53 (50%), Gaps = 5/53 (9%)
Query 128 EGGTLMECRISPKVDYLRLFAI----LNDQQEAVQRAILRLKPPRVFPGLTCW 176
+GG C++ K+D +++A+ N + E +A+ RL P + TCW
Sbjct 178 DGGYGFVCKVKHKIDD-KIYAVKRVEFNSEAEPEVKALARLDHPNIVRYFTCW 229
Lambda K H
0.327 0.142 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5364689396
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40