bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0063_orf2
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
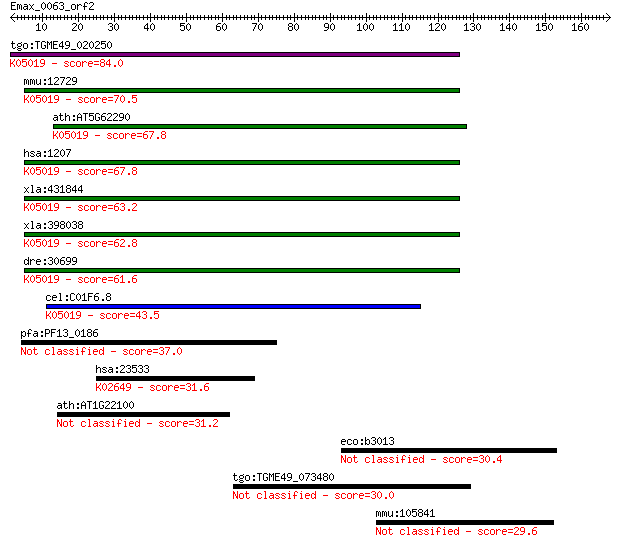
tgo:TGME49_020250 chloride channel, nucleotide-sensitive, 1A, ... 84.0 2e-16
mmu:12729 Clns1a, 2610036D06Rik, 2610100O04Rik, Clci, Clcni, I... 70.5 3e-12
ath:AT5G62290 nucleotide-sensitive chloride conductance regula... 67.8 2e-11
hsa:1207 CLNS1A, CLCI, CLNS1B, ICln; chloride channel, nucleot... 67.8 2e-11
xla:431844 clns1a, MGC81186, clci, clns1b, icln; chloride chan... 63.2 4e-10
xla:398038 clns1a, MGC85110, icln; regulatory protein; K05019 ... 62.8 5e-10
dre:30699 icln; swelling dependent chloride channel; K05019 ch... 61.6 1e-09
cel:C01F6.8 icl-1; ICLn ion channel homolog family member (icl... 43.5 3e-04
pfa:PF13_0186 conserved Plasmodium protein, unknown function 37.0 0.029
hsa:23533 PIK3R5, F730038I15Rik, FOAP-2, P101-PI3K, p101; phos... 31.6 1.2
ath:AT1G22100 ATP binding / inositol pentakisphosphate 2-kinase 31.2 1.6
eco:b3013 yqhG, ECK3005, JW5500, yqhF; conserved protein 30.4 2.5
tgo:TGME49_073480 axonemal beta dynein heavy chain, putative (... 30.0 3.8
mmu:105841 Dennd3, AI447457, E030003N15Rik; DENN/MADD domain c... 29.6 4.9
> tgo:TGME49_020250 chloride channel, nucleotide-sensitive, 1A,
putative ; K05019 chloride channel, nucleotide-sensitive,
1A
Length=176
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 70/133 (52%), Gaps = 31/133 (23%)
Query 1 ASIFISGKDAGRGELFITSQRVAWLSDVGAS--------FALNYVSIVLHALSTDPQACD 52
A + + G++ G G +ITS+R+AWL+ GA+ +++Y SIVLHALS DP +
Sbjct 35 AKLILQGENHGVGTFYITSRRIAWLAKPGAASGDEQRRDISVDYPSIVLHALSRDPNSGH 94
Query 53 RPCLYCQIKGEALADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQR 112
PC+YCQ+K +A A E +D + E++ VP L
Sbjct 95 EPCIYCQLKSDA-----------------------AAEDDEDYVIPEMRIVPTSSERLDS 131
Query 113 LFTVMSDMAALHP 125
LF +MS+MAAL+P
Sbjct 132 LFKIMSEMAALNP 144
> mmu:12729 Clns1a, 2610036D06Rik, 2610100O04Rik, Clci, Clcni,
ICLN; chloride channel, nucleotide-sensitive, 1A; K05019 chloride
channel, nucleotide-sensitive, 1A
Length=241
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 62/121 (51%), Gaps = 11/121 (9%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
++GK G G L+I R++WL G F+L Y +I LHA+S DP A + LY + +
Sbjct 32 LNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHAVSRDPNAYPQEHLYVMVNAK- 90
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
L + S P+S + D + + E +FVP D S L+ +FT M + ALH
Sbjct 91 LGEESK-EPLSDEDEEDN---------DDVEPISEFRFVPSDKSALEAMFTAMCECQALH 140
Query 125 P 125
P
Sbjct 141 P 141
> ath:AT5G62290 nucleotide-sensitive chloride conductance regulator
(ICln) family protein; K05019 chloride channel, nucleotide-sensitive,
1A
Length=228
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 62/117 (52%), Gaps = 16/117 (13%)
Query 13 GELFITSQRVAWLSDV--GASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEALADLSN 70
G L+ITS+++ WLSDV +A++++SI LHA+S DP+A PC+Y QI
Sbjct 49 GTLYITSRKLIWLSDVDMAKGYAVDFLSISLHAVSRDPEAYSSPCIYTQI---------- 98
Query 71 GNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALHPDP 127
+ E+ E D + E++ VP D + L+ LF V + A L+P+P
Sbjct 99 ----EVEEDEDDESDSESTEVLDLSKIREMRLVPSDSTQLETLFDVFCECAELNPEP 151
> hsa:1207 CLNS1A, CLCI, CLNS1B, ICln; chloride channel, nucleotide-sensitive,
1A; K05019 chloride channel, nucleotide-sensitive,
1A
Length=237
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 58/121 (47%), Gaps = 10/121 (8%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
++GK G G L+I R++WL G F+L Y +I LHALS D C LY + +
Sbjct 27 LNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHALSRDRSDCLGEHLYVMVNAK- 85
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
S + EE + D + + E +FVP D S L+ +FT M + ALH
Sbjct 86 ---------FEEESKEPVADEEEEDSDDDVEPITEFRFVPSDKSALEAMFTAMCECQALH 136
Query 125 P 125
P
Sbjct 137 P 137
> xla:431844 clns1a, MGC81186, clci, clns1b, icln; chloride channel,
nucleotide-sensitive, 1A; K05019 chloride channel, nucleotide-sensitive,
1A
Length=240
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 63/121 (52%), Gaps = 6/121 (4%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
+ G+ G G L+I R++WL+ G F+L Y SI LHA+S D A LY + +
Sbjct 25 VGGRGLGAGTLYIAESRLSWLNSSGLGFSLEYPSISLHAISRDTAAYPEEHLYVMVNSKL 84
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
G+ + + EE+E+ D++ + E++FVP + S L +F+ M D ALH
Sbjct 85 ------GDKEDKEAPMADQEEEESEDDDDEEPITEIRFVPAEKSDLGEMFSAMCDCQALH 138
Query 125 P 125
P
Sbjct 139 P 139
> xla:398038 clns1a, MGC85110, icln; regulatory protein; K05019
chloride channel, nucleotide-sensitive, 1A
Length=241
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 62/121 (51%), Gaps = 5/121 (4%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
+ G+ G G L+I R++WL+ G F+L Y SI LHA+S D A LY + +
Sbjct 25 VGGRGLGPGTLYIAESRLSWLNGSGLGFSLEYPSISLHAISRDTAAYPEEHLYVMVNSK- 83
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
LAD + + E ++ D++ + E++FVP + S L +F+ M D ALH
Sbjct 84 LADKED----KEAHMADQEEEESEDDDDDEEPITEIRFVPGEKSDLGEMFSAMCDCQALH 139
Query 125 P 125
P
Sbjct 140 P 140
> dre:30699 icln; swelling dependent chloride channel; K05019
chloride channel, nucleotide-sensitive, 1A
Length=249
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 59/121 (48%), Gaps = 2/121 (1%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
+ GK G G LF+ +++W G F L Y +I LHA+S D A LY + +
Sbjct 25 LDGKRLGLGTLFVAEAQLSWFDGSGMGFCLEYPTISLHAISRDLSAFPEEHLYVMVNAK- 83
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
L D P+ + E+++ + + E++FVP D + L+ +F+ M D ALH
Sbjct 84 LDDEGEAAPLEKDPDEEDENEEDSDSEGSGE-ITEIRFVPSDKAALEPMFSAMCDCQALH 142
Query 125 P 125
P
Sbjct 143 P 143
> cel:C01F6.8 icl-1; ICLn ion channel homolog family member (icl-1);
K05019 chloride channel, nucleotide-sensitive, 1A
Length=205
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 14/107 (13%)
Query 11 GRGELFITSQRVAWLSDVGAS--FALNYVSIVLHALSTDPQACDRPCLYCQI-KGEALAD 67
G G L+IT V W+S + F++ Y +IVLHA+STD ++ + + ++ +
Sbjct 30 GNGTLYITDSAVIWISSAAGTKGFSVAYPAIVLHAISTDVSVFPSEHIFVMVDQRKSGLE 89
Query 68 LSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLF 114
L+ S D P +E++FVPDD L +++
Sbjct 90 LAAAELEDEESDDDEEEP-----------ALEIRFVPDDKDSLSQIY 125
> pfa:PF13_0186 conserved Plasmodium protein, unknown function
Length=234
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 25/110 (22%), Positives = 45/110 (40%), Gaps = 39/110 (35%)
Query 4 FISGK-DAGRGELFITSQRVAWLSD------------------------VGASFALNYVS 38
FI K + G G+L+I +R+ W+++ +F L+ ++
Sbjct 33 FIYNKLNLGEGKLYILEKRLLWINENANKKNVTNFKELCTNNIYLNHYEKNRNFYLHLLN 92
Query 39 -----------IVLHALSTDPQACDRPCLYCQIK---GEALADLSNGNPV 74
I LHA+++D + CD C+Y Q+ + L + N PV
Sbjct 93 EVNNISIDSSNIALHAITSDKKICDNSCVYIQLNTDISDHLENWKNDEPV 142
> hsa:23533 PIK3R5, F730038I15Rik, FOAP-2, P101-PI3K, p101; phosphoinositide-3-kinase,
regulatory subunit 5; K02649 phosphoinositide-3-kinase,
regulatory subunit
Length=880
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Query 25 LSDVGASFALNYVSIVLHAL-STDPQACDRPCLYCQIKGEALADL 68
LS G S Y +++LHA +T CD P L+C+++ + LA+L
Sbjct 182 LSTPGHSPHSAYTTLLLHAFQATFGAHCDVPGLHCRLQAKTLAEL 226
> ath:AT1G22100 ATP binding / inositol pentakisphosphate 2-kinase
Length=441
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 4/51 (7%)
Query 14 ELFITSQRVAWLSDVGASFALNYVSIVLH---ALSTDPQACDRPCLYCQIK 61
E ITSQR +W +DV AS N S++L L D+PCL +IK
Sbjct 118 EKIITSQRPSWRADV-ASVDTNRSSVLLMDDLTLFAHGHVEDKPCLSVEIK 167
> eco:b3013 yqhG, ECK3005, JW5500, yqhF; conserved protein
Length=308
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 34/69 (49%), Gaps = 9/69 (13%)
Query 93 DDDAMVELKFVPDDPSCL--QRLFTVMSDMAALHPDPDSA------TLDGDEDELFDEAS 144
D+D +E +F+ D+P C + LF+ + ++ L DSA TL +E +
Sbjct 215 DEDMQIERQFIYDNPICFGEESLFSRVDEIRVLEKTADSARIHVRFTLTNGNNEEQELVL 274
Query 145 LRRQG-WEF 152
RR+G WE
Sbjct 275 QRREGKWEI 283
> tgo:TGME49_073480 axonemal beta dynein heavy chain, putative
(EC:5.99.1.3)
Length=4273
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 30/75 (40%), Gaps = 16/75 (21%)
Query 63 EALADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFT------- 115
EAL + + P S+DS ADD K + D LQRLFT
Sbjct 2911 EALCKMFSIKPAKVKSADSM-------RKADDYWTASKKHLLSDTKFLQRLFTYDKDHIP 2963
Query 116 --VMSDMAALHPDPD 128
VMSD+ DPD
Sbjct 2964 VEVMSDILPYQTDPD 2978
> mmu:105841 Dennd3, AI447457, E030003N15Rik; DENN/MADD domain
containing 3
Length=1274
Score = 29.6 bits (65), Expect = 4.9, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 21/49 (42%), Gaps = 2/49 (4%)
Query 103 VPDDPSCLQRLFTVMSDMAALHPDPDSATLDGDEDELFDEASLRRQGWE 151
+PD P L + F LHPD A L D +E RR+ W+
Sbjct 387 IPDVPLLLAQTFIQRVQSLQLHPDLHLAHLSASTD--LNEGRARRRAWQ 433
Lambda K H
0.315 0.132 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40