bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0042_orf2
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
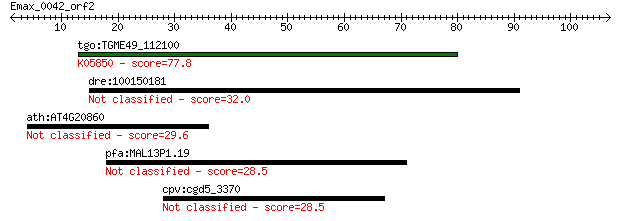
tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transp... 77.8 7e-15
dre:100150181 tmtc3; transmembrane and tetratricopeptide repea... 32.0 0.46
ath:AT4G20860 FAD-binding domain-containing protein 29.6 2.6
pfa:MAL13P1.19 peptidase, putative 28.5 4.8
cpv:cgd5_3370 hypothetical protein 28.5 5.3
> tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1822
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 51/67 (76%), Gaps = 0/67 (0%)
Query 13 PQCFPRITRMFPEFGMREEDLSEKASLALGLRGRSPSRMNDRCMISTLAVAKKYARRAKV 72
P+ FP + ++ PEFG REE L E SLAL LRGR+P RMN+R +++ AKKYA++A++
Sbjct 1699 PRVFPSLYKLAPEFGSREETLEETHSLALELRGRTPERMNERLGLASYTFAKKYAKKARL 1758
Query 73 LTISRMK 79
+T+SR++
Sbjct 1759 MTLSRLR 1765
> dre:100150181 tmtc3; transmembrane and tetratricopeptide repeat
containing 3
Length=906
Score = 32.0 bits (71), Expect = 0.46, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 38/78 (48%), Gaps = 11/78 (14%)
Query 15 CFPRITRMFPEFGMREEDLSEKASLALGLRGRSPSRMNDRCMISTLAVA--KKYARRAKV 72
CF RI M P ++ K +L + + +RC++ TLA+A ++Y RR
Sbjct 759 CFERILSMDPS------NVQAKHNLCVVYFEERELQRAERCLVDTLAMAPQEEYIRRHLA 812
Query 73 LTISRMKARRGASRAGQP 90
+ ++M A S AGQP
Sbjct 813 IVRNKMAA---MSAAGQP 827
> ath:AT4G20860 FAD-binding domain-containing protein
Length=530
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 4 FVGLLFARIPQCFPRITRMFPEFGMREEDLSE 35
F L I + P + + FPE G+R +D SE
Sbjct 307 FQTLFLGGIDRLIPLMNQKFPELGLRSQDCSE 338
> pfa:MAL13P1.19 peptidase, putative
Length=9271
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Query 18 RITRMFPEFGMREEDLSEKASLALGLRGRSPSRMNDRCMISTLAVAKKYARRA 70
+IT MFPE+ ++++ +EK ++L + +S +N R S+ ++ K R+
Sbjct 3662 KITDMFPEYKNKKDNHNEK--MSLNILAQSYPVVNKRKYKSSTSLNNKRGRKG 3712
> cpv:cgd5_3370 hypothetical protein
Length=1019
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Query 28 MREEDLSEKASLALGLRGRSPSRMNDRCMIS--TLAVAKKY 66
+ +EDLS+K + L R +++ND+C+I+ TL ++ Y
Sbjct 634 IHKEDLSQKICIYQSLLQRIINKLNDKCLINKETLPLSSLY 674
Lambda K H
0.326 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40