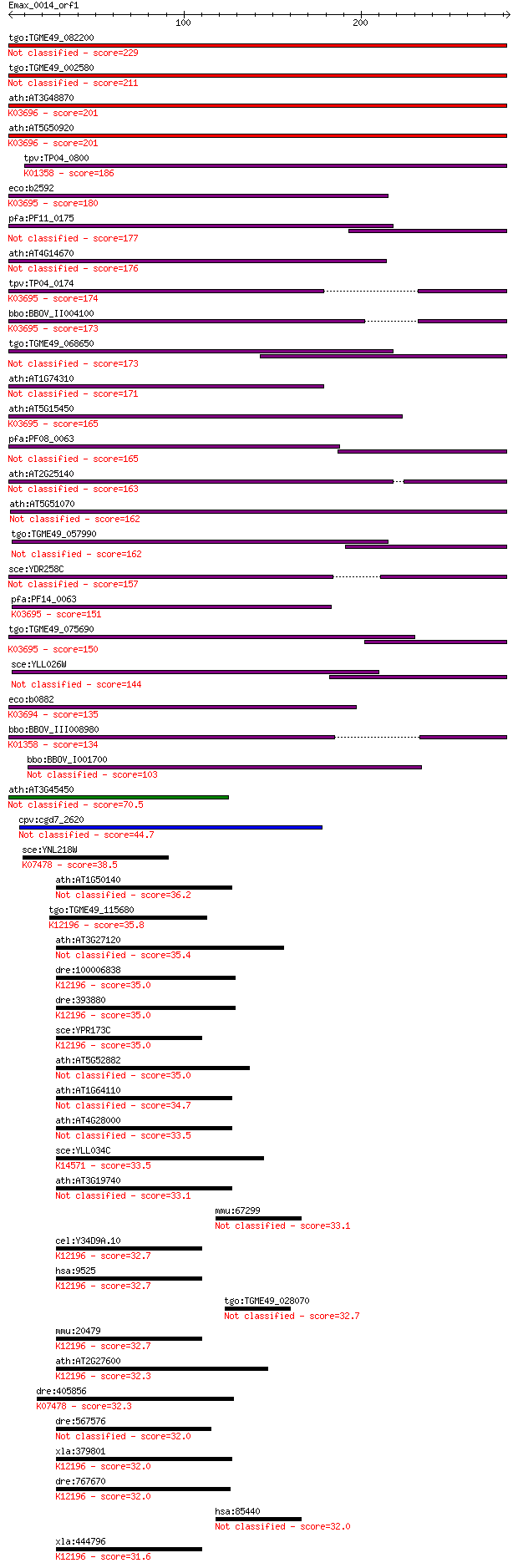
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0014_orf1
Length=283
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082200 clpB protein, putative 229 1e-59
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 211 2e-54
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 201 2e-51
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 201 2e-51
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 186 6e-47
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 180 5e-45
pfa:PF11_0175 heat shock protein 101, putative 177 3e-44
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 176 1e-43
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 174 3e-43
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 173 7e-43
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 173 8e-43
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 171 2e-42
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 165 2e-40
pfa:PF08_0063 ClpB protein, putative 165 2e-40
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 163 7e-40
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 162 1e-39
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 162 2e-39
sce:YDR258C HSP78; Hsp78p 157 5e-38
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 151 2e-36
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 150 4e-36
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 144 4e-34
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 135 2e-31
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 134 3e-31
bbo:BBOV_I001700 19.m02115; chaperone clpB 103 8e-22
ath:AT3G45450 Clp amino terminal domain-containing protein 70.5 7e-12
cpv:cgd7_2620 ClpB ATpase (bacterial), signal peptide 44.7 4e-04
sce:YNL218W MGS1; Mgs1p; K07478 putative ATPase 38.5 0.032
ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/... 36.2 0.16
tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12... 35.8 0.18
ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/... 35.4 0.23
dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog ... 35.0 0.30
dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sortin... 35.0 0.31
sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-A... 35.0 0.31
ath:AT5G52882 ATP binding / nucleoside-triphosphatase/ nucleot... 35.0 0.33
ath:AT1G64110 AAA-type ATPase family protein 34.7 0.43
ath:AT4G28000 ATP binding / ATPase/ nucleoside-triphosphatase/... 33.5 0.88
sce:YLL034C RIX7; Putative ATPase of the AAA family, required ... 33.5 1.0
ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/... 33.1 1.1
mmu:67299 Dock7, 3110056M06Rik, Gm430, m, mKIAA1771; dedicator... 33.1 1.2
cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting... 32.7 1.4
hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting ... 32.7 1.4
tgo:TGME49_028070 hypothetical protein 32.7 1.5
mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting... 32.7 1.5
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 32.3 2.0
dre:405856 MGC85976; zgc:85976; K07478 putative ATPase 32.3 2.3
dre:567576 spectrin repeat containing, nuclear envelope 1-like 32.0 2.4
xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog... 32.0 2.5
dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sort... 32.0 2.5
hsa:85440 DOCK7, KIAA1771, ZIR2; dedicator of cytokinesis 7 32.0
xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-a... 31.6 3.2
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 229 bits (584), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 128/301 (42%), Positives = 187/301 (62%), Gaps = 21/301 (6%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VPK L + + ++DL L AG + RGEFEKRMK +I + ++ VILF+DE+H L+G
Sbjct 302 GMVPKSLENKILFAVDLGALIAGATYRGEFEKRMKALIRYAVNQEGRVILFIDELHMLMG 361
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AGK+ G+ DA +LK P+ARGEI LVGATT EEYK+ IEKDAA RR + I +E PS +R
Sbjct 362 AGKSDGTMDAANLLKPPMARGEIRLVGATTQEEYKI-IEKDAAMERRLKPIFIEEPSTDR 420
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVS- 179
A+ IL+K+ +E H +++ ++ + V +S +Y + R PDKA+DLLDEA + K+V
Sbjct 421 AIYILRKLSDKFESHHEMKISDEAIVAAVMLSHKYIRNRKLPDKAIDLLDEAAATKRVKW 480
Query 180 -----HNKKLVLLKMQIADMESNAADATAEDLQKLKEE--------------LAALEALT 220
++ ++ + + E +D Q+++++ E
Sbjct 481 DLRSLNDDEVAMERKNSEQFEEETRKVEQQDQQRIEKQNHGDGTEEELKNLEKEIEEQSL 540
Query 221 ADGRRLVLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADA 280
+ + LVL DVA ++ LWTGIPL K+T+DE S++LRL+D+L RVIGQ AVKAVADA
Sbjct 541 HEKQELVLTADDVAQVISLWTGIPLAKLTDDEKSKILRLSDLLHSRVIGQDDAVKAVADA 600
Query 281 L 281
+
Sbjct 601 M 601
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 211 bits (538), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 124/295 (42%), Positives = 183/295 (62%), Gaps = 21/295 (7%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L G ++ SLD+ +L +G+SMRGEFE+RMK I+ +L + ILF+DEIHTL+G
Sbjct 327 GDVPDALLGVQVFSLDVGSLLSGSSMRGEFERRMKGILDYLFASDRSTILFIDEIHTLMG 386
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AGK G DA +LK LARG + ++GATT EY+ HIE+D AF RRF I ++ P +
Sbjct 387 AGKADGPMDAANLLKPALARGALRVIGATTRAEYRKHIERDMAFARRFVTIEMKEPDVAK 446
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+++LK ++ + E H + + + L +S +Y K R PDKA+DL+D+AC+ KKV
Sbjct 447 TITMLKGIRKNLENHHKLTITDGALVAAATLSDRYIKSRQLPDKAIDLIDDACAIKKVKS 506
Query 181 NKKLVLLKMQIADMESN----AADATA----------EDLQKLKEELAALEALTADGRRL 226
++ L+ + A+ E N + DA++ +DL+KL+ E+A EA AD
Sbjct 507 LRR--LMNERAANEEKNDNGGSGDASSSANGDGEKKRDDLEKLEAEIAK-EAEDAD---- 559
Query 227 VLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
L DVA ++ L TGIP+ K+TE + R+L L L +VIGQ +AV+AVA+A+
Sbjct 560 ALTDADVADVVSLRTGIPVNKLTETDRQRLLSLPASLQAQVIGQEEAVEAVANAI 614
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 201 bits (512), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 118/305 (38%), Positives = 178/305 (58%), Gaps = 26/305 (8%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ + G+ +I+LD+ L AGT RGEFE+R+K+++ + R+ ++ILF+DE+HTL+G
Sbjct 343 GDVPETIEGKTVITLDMGLLVAGTKYRGEFEERLKKLMEEI-RQSDEIILFIDEVHTLIG 401
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA ILK LARGE+ +GATT++EY+ HIEKD A RRFQ + V P+ E
Sbjct 402 AGAAEGAIDAANILKPALARGELQCIGATTIDEYRKHIEKDPALERRFQPVKVPEPTVEE 461
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
A+ IL+ ++ YE H + ++ L +S QY R PDKA+DL+DEA S ++ H
Sbjct 462 AIQILQGLRERYEIHHKLRYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEAGSRVRLRH 521
Query 181 ------------------NKKLVLLKMQIADMESNAADATAE------DLQKLKEELAAL 216
+K ++ Q +M + D E ++ +E+A
Sbjct 522 AQLPEEARELEKQLRQITKEKNEAVRSQDFEMAGSHRDREIELKAEIANVLSRGKEVAKA 581
Query 217 EALTADGRRLVLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKA 276
E +G V E D+ HI+ WTGIP+ K++ DE SR+L++ L RVIGQ +AVKA
Sbjct 582 ENEAEEGGPTVTE-SDIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTRVIGQDEAVKA 640
Query 277 VADAL 281
++ A+
Sbjct 641 ISRAI 645
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 201 bits (511), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 117/306 (38%), Positives = 184/306 (60%), Gaps = 28/306 (9%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ + G+++I+LD+ L AGT RGEFE+R+K+++ + R+ ++ILF+DE+HTL+G
Sbjct 322 GDVPETIEGKKVITLDMGLLVAGTKYRGEFEERLKKLMEEI-RQSDEIILFIDEVHTLIG 380
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA ILK LARGE+ +GATTL+EY+ HIEKD A RRFQ + V P+ +
Sbjct 381 AGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVKVPEPTVDE 440
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+ ILK ++ YE H + ++ L +S QY R PDKA+DL+DEA S ++ H
Sbjct 441 TIQILKGLRERYEIHHKLRYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEAGSRVRLRH 500
Query 181 NK-----KLVLLKMQIADMESNAADATAEDLQK----------LKEELAAL--------- 216
+ + + +++ E N A +D +K L+ E++A+
Sbjct 501 AQVPEEARELEKELRQITKEKNEA-VRGQDFEKAGTLRDREIELRAEVSAIQAKGKEMSK 559
Query 217 -EALTADGRRLVLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVK 275
E+ T + +V E D+ HI+ WTGIP+ K++ DE R+L++ + L R+IGQ +AVK
Sbjct 560 AESETGEEGPMVTE-SDIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKRIIGQDEAVK 618
Query 276 AVADAL 281
A++ A+
Sbjct 619 AISRAI 624
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 186 bits (473), Expect = 6e-47, Method: Compositional matrix adjust.
Identities = 106/272 (38%), Positives = 159/272 (58%), Gaps = 6/272 (2%)
Query 10 RRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSD 69
+RI+ L L AGT RG+FE+R+ ++I + + D+IL +DE H L+G G GS D
Sbjct 324 KRILQLQFGLLIAGTKFRGQFEERLTKLIDEI-KSAGDIILVIDEAHMLIGGGAGDGSID 382
Query 70 ATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVK 129
A +LK PL+RGEI + TT +EYK + EKD A RRF I V+ PS E L IL +
Sbjct 383 AANLLKPPLSRGEIQCIAITTPKEYKKYFEKDMALSRRFHPIYVDEPSDEDTLKILNGIS 442
Query 130 HHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSHNKKLVLLKM 189
Y +FHGVE +D ++L + S QY R PDKA+D++DE+ S+ K+ + +L K
Sbjct 443 SSYGEFHGVEYTQDSIKLALKYSKQYINDRFLPDKAIDIMDESGSFAKIQYQNELKREKN 502
Query 190 QIADMESNAADATAEDLQKLKEELAALEALTADGRRLVLELHDVAHILHLWTGIPLGKMT 249
+I D SN+ E+ K+ E+ + + + G+ ++ VA ++ +WTGIPL K+T
Sbjct 503 EIPD-PSNSTSPEGENETKV-EQKSEKKDQSVLGQ---VKPEHVAEVMSIWTGIPLKKLT 557
Query 250 EDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
E+ + + + L VIGQ +AVK V A+
Sbjct 558 RGEMDIIRNMEEDLHKMVIGQEEAVKNVCKAI 589
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 180 bits (457), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 94/224 (41%), Positives = 145/224 (64%), Gaps = 10/224 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+GL+GRR+++LD+ L AG RGEFE+R+K ++ L +++ +VILF+DE+HT+VG
Sbjct 226 GEVPEGLKGRRVLALDMGALVAGAKYRGEFEERLKGVLNDLAKQEGNVILFIDELHTMVG 285
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AGK G+ DA +LK LARGE+ VGATTL+EY+ +IEKDAA RRFQ + V PS E
Sbjct 286 AGKADGAMDAGNMLKPALARGELHCVGATTLDEYRQYIEKDAALERRFQKVFVAEPSVED 345
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVS- 179
++IL+ +K YE H V++ + + +S +Y R PDKA+DL+DEA S ++
Sbjct 346 TIAILRGLKERYELHHHVQITDPAIVAAATLSHRYIADRQLPDKAIDLIDEAASSIRMQI 405
Query 180 ---------HNKKLVLLKMQIADMESNAADATAEDLQKLKEELA 214
+++++ LK++ + + +A+ + L L EEL+
Sbjct 406 DSKPEELDRLDRRIIQLKLEQQALMKESDEASKKRLDMLNEELS 449
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 177 bits (450), Expect = 3e-44, Method: Composition-based stats.
Identities = 95/217 (43%), Positives = 138/217 (63%), Gaps = 12/217 (5%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VPK L+G +ISL+ ++GTS RGEFE RMK II L+ +K +ILFVDEIH L+G
Sbjct 255 GDVPKELQGYTVISLNFRKFTSGTSYRGEFETRMKNIIKELKNKKNKIILFVDEIHLLLG 314
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AGK G +DA +LK L++GEI L+GATT+ EY+ IE +AF RRF+ I+VE PS +
Sbjct 315 AGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFERRFEKILVEPPSVDM 374
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+ IL+ +K YE F+G+ + + L +S ++ K R PDKA+DLL++ACS+ +V
Sbjct 375 TVKILRSLKSKYENFYGINITDKALVAAAKISDRFIKDRYLPDKAIDLLNKACSFLQVQL 434
Query 181 NKKLVLLKMQIADMESNAADATAEDLQKLKEELAALE 217
+ K ++ D T D+++L E++ LE
Sbjct 435 SGKPRII------------DVTERDIERLSYEISTLE 459
Score = 36.6 bits (83), Expect = 0.099, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 193 DMESNAADATAEDLQKLKEELAALEALTADGRRL--VLELHDVAHILHLWTGIPLGKMTE 250
+++++ +A + L+ KE +A +EA T + + V + H V++I +G+PLG ++
Sbjct 528 ELQNSLKEAQQKYLELYKETVAYVEAKTHNAMNVDAVYQEH-VSYIYLRDSGMPLGSLSF 586
Query 251 DEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
+ L+L + LS +IG +K+++DA+
Sbjct 587 ESSKGALKLYNSLSKSIIGNEDIIKSLSDAV 617
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 176 bits (445), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 94/224 (41%), Positives = 143/224 (63%), Gaps = 11/224 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L G ++ISL+ + AGT++RG+FE+R+K ++ ++ + V+LF+DEIH +G
Sbjct 192 GDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKVVLFIDEIHMALG 251
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
A K SGS+DA ++LK LARG++ +GATTLEEY+ H+EKDAAF RRFQ + V PS
Sbjct 252 ACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERRFQQVFVAEPSVPD 311
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKV-- 178
+SIL+ +K YE HGV + + L L +S +Y R PDKA+DL+DE+C+ K
Sbjct 312 TISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKAIDLVDESCAHVKAQL 371
Query 179 --------SHNKKLVLLKMQIADMESNAADATAE-DLQKLKEEL 213
S +K++ L+++I +E D +E L ++++EL
Sbjct 372 DIQPEEIDSLERKVMQLEIEIHALEKEKDDKASEARLSEVRKEL 415
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 174 bits (441), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 85/178 (47%), Positives = 124/178 (69%), Gaps = 0/178 (0%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L+ R+++SLD+ + AGT RGEFE+R+KEI++ ++ + ++++F+DEIHTLVG
Sbjct 328 GDVPDSLKNRKVLSLDIAAIVAGTMYRGEFEERLKEILSEIENSQGEIVMFIDEIHTLVG 387
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG++ GS DA ILK LARGE+ +GATTL+EY+ IEKD A RRFQ I ++ P+ E
Sbjct 388 AGESQGSLDAGNILKPMLARGELRCIGATTLQEYRQKIEKDKALERRFQPIYIDEPNIEE 447
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKV 178
++IL+ +K YE HGV + + L V +S +Y R PDKA+DL+DEA + K+
Sbjct 448 TINILRGLKERYEVHHGVRILDSTLIQAVLLSNRYITDRYLPDKAIDLIDEAAAKLKI 505
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 232 DVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
D+A+++ WTGIPL K+ + + ++L+L D L R+IGQ +A+ AV +A+
Sbjct 661 DIANVVSKWTGIPLNKLIKSQKEKILQLNDELHKRIIGQQEAIDAVVNAV 710
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 173 bits (438), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 92/211 (43%), Positives = 132/211 (62%), Gaps = 10/211 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L+ R+ISLDL ++ AG+ RGEFE+R+K I+ +Q + ++I+F+DEIHT+VG
Sbjct 268 GDVPDSLKNTRVISLDLASMLAGSQYRGEFEERLKNILKEVQDSQGEIIMFIDEIHTVVG 327
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA ILK LARGE+ +GATTL+EY+ IEKD A RRFQ + V+ PS E
Sbjct 328 AGDAQGAMDAGNILKPMLARGELRCIGATTLQEYRQRIEKDKALERRFQPVYVDQPSVEE 387
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+SIL+ ++ YE HGV + + L +S +Y R PDKA+DL+DEA + K+
Sbjct 388 TISILRGLRERYEVHHGVRILDSALVEAAQLSDRYITDRFLPDKAIDLVDEAAARLKIQL 447
Query 181 NKK----------LVLLKMQIADMESNAADA 201
+ K L+ L+M+ + S+A D
Sbjct 448 SSKPIQLDGLERRLIQLEMERISISSDATDG 478
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 232 DVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
D+A ++ WTGIP+ K+ + ++L + D L R+IGQ +AV V A+
Sbjct 606 DIASVVSRWTGIPVNKLIRSQRDKILHIGDELRKRIIGQDEAVDIVTRAV 655
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 173 bits (438), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 102/232 (43%), Positives = 144/232 (62%), Gaps = 15/232 (6%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L GR++ISLDL L AG +RGEFE+R+K +I +Q +ILF+DEIH +VG
Sbjct 243 GDVPDTLAGRQLISLDLGALLAGAKLRGEFEERLKSVIREVQESSGQIILFIDEIHMVVG 302
Query 61 AGKTSGSS-DATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKE 119
AG S DA ILK LARGE+ +GATTL+EY+ +IEKD A RRFQ ++V+ P E
Sbjct 303 AGSAGESGMDAGNILKPMLARGELRCIGATTLDEYRKYIEKDKALERRFQVVLVDEPRVE 362
Query 120 RALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVS 179
ALSIL+ +K YE HGV + + L +S +Y + R PDKA+DL+DEA S K+
Sbjct 363 DALSILRGLKERYEMHHGVSIRDSALVAACVLSNRYIQDRFLPDKAIDLIDEAASKIKIE 422
Query 180 H----------NKKLVLLKMQ----IADMESNAADATAEDLQKLKEELAALE 217
++KL+ L+M+ ++DM+ A + LQ L++++A L+
Sbjct 423 VTSKPTRLDEIDRKLMQLEMEKISIVSDMKGGQDAAEQQRLQTLEKKMADLK 474
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 76/149 (51%), Gaps = 20/149 (13%)
Query 143 DVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSHNKKLVLLKMQIADMESN---AA 199
D+ E ++A ++++R+ +K DL E K ++ Q A+ E N AA
Sbjct 472 DLKEEQSRLNAIWEQERAEIEKIADLKQEIDEAK----------VEQQKAEREYNLNKAA 521
Query 200 DATAEDLQKLKEELAALEALT-----ADGR--RLVLELHDVAHILHLWTGIPLGKMTEDE 252
+ +L ++LA LEA + R R + D+A ++ WTGIP+ ++ E E
Sbjct 522 QIRYGKIPELTQKLATLEAAAKREELSSSRLLRDTVTAEDIAQVVGSWTGIPVSRLVEGE 581
Query 253 VSRVLRLADILSLRVIGQHQAVKAVADAL 281
++L L L+ RVIGQ + V++VA+A+
Sbjct 582 REKLLGLEKALNDRVIGQEEGVRSVAEAI 610
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 171 bits (434), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 87/178 (48%), Positives = 118/178 (66%), Gaps = 0/178 (0%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L R+ISLD+ L AG RGEFE+R+K ++ ++ + VILF+DEIH ++G
Sbjct 227 GDVPNSLTDVRLISLDMGALVAGAKYRGEFEERLKSVLKEVEDAEGKVILFIDEIHLVLG 286
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AGKT GS DA + K LARG++ +GATTLEEY+ ++EKDAAF RRFQ + V PS
Sbjct 287 AGKTEGSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPD 346
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKV 178
+SIL+ +K YE HGV + + L +SA+Y R PDKA+DL+DEAC+ +V
Sbjct 347 TISILRGLKEKYEGHHGVRIQDRALINAAQLSARYITGRHLPDKAIDLVDEACANVRV 404
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 165 bits (417), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 92/232 (39%), Positives = 140/232 (60%), Gaps = 10/232 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ L R++ISLD+ L AG RGEFE R+K ++ + + +ILF+DEIHT+VG
Sbjct 302 GDVPQALMNRKLISLDMGALIAGAKYRGEFEDRLKAVLKEVTDSEGQIILFIDEIHTVVG 361
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG T+G+ DA +LK L RGE+ +GATTL+EY+ +IEKD A RRFQ + V+ P+ E
Sbjct 362 AGATNGAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALERRFQQVYVDQPTVED 421
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+SIL+ ++ YE HGV + + L +S +Y R PDKA+DL+DEA + K+
Sbjct 422 TISILRGLRERYELHHGVRISDSALVEAAILSDRYISGRFLPDKAIDLVDEAAAKLKMEI 481
Query 181 NKK----------LVLLKMQIADMESNAADATAEDLQKLKEELAALEALTAD 222
K ++ L+M+ + ++ A+ E L +++ EL L+ A+
Sbjct 482 TSKPTALDELDRSVIKLEMERLSLTNDTDKASRERLNRIETELVLLKEKQAE 533
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 165 bits (417), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 88/188 (46%), Positives = 124/188 (65%), Gaps = 1/188 (0%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L+GR+++SLD+ +L AG RG+FE+R+K I+ +Q + V++F+DEIHT+VG
Sbjct 379 GDVPDSLKGRKLVSLDMSSLIAGAKYRGDFEERLKSILKEVQDAEGQVVMFIDEIHTVVG 438
Query 61 AGKTS-GSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKE 119
AG + G+ DA ILK LARGE+ +GATT+ EY+ IEKD A RRFQ I+VE PS +
Sbjct 439 AGAVAEGALDAGNILKPMLARGELRCIGATTVSEYRQFIEKDKALERRFQQILVEQPSVD 498
Query 120 RALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVS 179
+SIL+ +K YE HGV + + L +S +Y R PDKA+DL+DEA S K+
Sbjct 499 ETISILRGLKERYEVHHGVRILDSALVQAAVLSDRYISYRFLPDKAIDLIDEAASNLKIQ 558
Query 180 HNKKLVLL 187
+ K + L
Sbjct 559 LSSKPIQL 566
Score = 39.3 bits (90), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 15/95 (15%)
Query 187 LKMQIADMESNAADATAEDLQKLKEELAALEALTADGRRLVLELHDVAHILHLWTGIPLG 246
L+ Q+ E N + E + LK+E+ + D+ +I+ + TGI L
Sbjct 716 LEKQLKKAEENYLNDIPEKSRILKDEVTS---------------EDIVNIVSMSTGIRLN 760
Query 247 KMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
K+ + E ++L L + L ++IGQ AVK V A+
Sbjct 761 KLLKSEKEKILNLENELHKQIIGQDDAVKVVTKAV 795
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 163 bits (412), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 89/227 (39%), Positives = 139/227 (61%), Gaps = 10/227 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ L R++ISLD+ +L AG RG+FE+R+K ++ + ILF+DEIHT+VG
Sbjct 307 GDVPEPLMNRKLISLDMGSLLAGAKFRGDFEERLKAVMKEVSASNGQTILFIDEIHTVVG 366
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA+ +LK L RGE+ +GATTL EY+ +IEKD A RRFQ ++ PS E
Sbjct 367 AGAMDGAMDASNLLKPMLGRGELRCIGATTLTEYRKYIEKDPALERRFQQVLCVQPSVED 426
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+SIL+ ++ YE HGV + + L ++ +Y +R PDKA+DL+DEA + K+
Sbjct 427 TISILRGLRERYELHHGVTISDSALVSAAVLADRYITERFLPDKAIDLVDEAGAKLKMEI 486
Query 181 NKK----------LVLLKMQIADMESNAADATAEDLQKLKEELAALE 217
K ++ L+M+ ++++ A+ E LQK++ +L+ L+
Sbjct 487 TSKPTELDGIDRAVIKLEMEKLSLKNDTDKASKERLQKIENDLSTLK 533
Score = 54.3 bits (129), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 224 RRLVLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
R +V +L D+A I+ WTGIPL + + E +++ L ++L RVIGQ AVK+VADA+
Sbjct 613 REVVTDL-DIAEIVSKWTGIPLSNLQQSEREKLVMLEEVLHHRVIGQDMAVKSVADAI 669
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 102/309 (33%), Positives = 169/309 (54%), Gaps = 31/309 (10%)
Query 2 AVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGA 61
+ P L +RI+SLD+ L AG RGE E R+ +I+ +++ K VILF+DE+HTL+G+
Sbjct 337 SAPGFLLTKRIMSLDIGLLMAGAKERGELEARVTALISEVKKSGK-VILFIDEVHTLIGS 395
Query 62 GKTS----GSS-DATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAP 116
G GS D +LK L RGE+ + +TTL+E++ EKD A RRFQ +++ P
Sbjct 396 GTVGRGNKGSGLDIANLLKPSLGRGELQCIASTTLDEFRSQFEKDKALARRFQPVLINEP 455
Query 117 SKERALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWK 176
S+E A+ IL ++ YE H + + ++ V +S++Y R PDKA+DL+DEA S
Sbjct 456 SEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIADRFLPDKAIDLIDEAGSRA 515
Query 177 KVSHNKK-----LVLLKMQIADM-------------------ESNAADATAEDLQKLKEE 212
++ +K + +L D + + DA +++ +L EE
Sbjct 516 RIEAFRKKKEDAICILSKPPNDYWQEIKTVQAMHEVVLSSRQKQDDGDAISDESGELVEE 575
Query 213 LAALEALTADGRRLVLELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQ 272
++L D +++ D+A + +W+GIP+ ++T DE ++ L D L RV+GQ +
Sbjct 576 -SSLPPAAGDDEPILVGPDDIAAVASVWSGIPVQQITADERMLLMSLEDQLRGRVVGQDE 634
Query 273 AVKAVADAL 281
AV A++ A+
Sbjct 635 AVAAISRAV 643
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 162 bits (409), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 90/222 (40%), Positives = 136/222 (61%), Gaps = 11/222 (4%)
Query 3 VPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAG 62
VP LR R ++SLD+ +L AG RGEFE+R+ ++ ++ +ILF+DEIH ++GAG
Sbjct 227 VPSNLRCR-LVSLDVGSLIAGAKFRGEFEERLTAVLQEVKDAAGKIILFIDEIHVILGAG 285
Query 63 KTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERAL 122
KT G+ DA +LK LARGE+ +GATTL+EY+ ++EKDAAF RRFQ + V PS + +
Sbjct 286 KTEGALDAANLLKPMLARGELRCIGATTLDEYRKYVEKDAAFERRFQQVHVREPSVQATI 345
Query 123 SILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSHNK 182
SIL+ +K Y HGV + + L ++ +Y R PDKA+DL+DEAC+ +V +
Sbjct 346 SILRGLKDRYASHHGVRILDSALVEAAQLADRYITSRFLPDKAIDLMDEACAIARVQVDS 405
Query 183 KL----------VLLKMQIADMESNAADATAEDLQKLKEELA 214
K V L++++ +E A+ + L ++KE L
Sbjct 406 KPEAVDVLERQKVQLEVELLALEKEKDPASQKRLAEVKEHLG 447
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 0/91 (0%)
Query 191 IADMESNAADATAEDLQKLKEELAALEALTADGRRLVLELHDVAHILHLWTGIPLGKMTE 250
+A++ +A +KL+EE E V+ +A ++H WT IP+ K+T+
Sbjct 496 VAELRFDALPGVEARFKKLQEEQEEYERTHKPLLTEVVGPEQIADVVHRWTNIPVQKLTQ 555
Query 251 DEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
E R L L L+ +VIGQ QAV+AV A+
Sbjct 556 TETERFLTLGKSLAEQVIGQPQAVEAVTQAI 586
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 157 bits (396), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 82/183 (44%), Positives = 119/183 (65%), Gaps = 2/183 (1%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L+ + +++LDL +L AG RGEFE+R+K+++ + + VI+F+DE+H L+G
Sbjct 163 GEVPDSLKDKDLVALDLGSLIAGAKYRGEFEERLKKVLEEIDKANGKVIVFIDEVHMLLG 222
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
GKT GS DA+ ILK LARG + + ATTL+E+K+ IEKD A RRFQ I++ PS
Sbjct 223 LGKTDGSMDASNILKPKLARG-LRCISATTLDEFKI-IEKDPALSRRFQPILLNEPSVSD 280
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
+SIL+ +K YE HGV + + L +S +Y R PDKA+DL+DEAC+ ++ H
Sbjct 281 TISILRGLKERYEVHHGVRITDTALVSAAVLSNRYITDRFLPDKAIDLVDEACAVLRLQH 340
Query 181 NKK 183
K
Sbjct 341 ESK 343
Score = 38.9 bits (89), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 7/77 (9%)
Query 211 EELAALEALTADGRRLVLELHD------VAHILHLWTGIPLGKMTEDEVSRVLRLADILS 264
E+ AL + DG ++ L LHD ++ ++ TGIP + + + R+L + + L
Sbjct 445 EKKVALSEKSKDGDKVNL-LHDSVTSDDISKVVAKMTGIPTETVMKGDKDRLLYMENSLK 503
Query 265 LRVIGQHQAVKAVADAL 281
RV+GQ +A+ A++DA+
Sbjct 504 ERVVGQDEAIAAISDAV 520
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 151 bits (382), Expect = 2e-36, Method: Composition-based stats.
Identities = 74/180 (41%), Positives = 117/180 (65%), Gaps = 1/180 (0%)
Query 3 VPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAG 62
VP L+ RI L+L N+ AGT RGEFE++MK +++ + ++KK+ ILF+DEIH +VGAG
Sbjct 573 VPYHLKNCRIFQLNLGNIVAGTKYRGEFEEKMKHLLSNMNKKKKN-ILFIDEIHVIVGAG 631
Query 63 KTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERAL 122
GS DA+ +LK L+ + +G TT +EY IE D A RRF + + + +
Sbjct 632 SGEGSLDASNLLKPFLSSDNLQCIGTTTFQEYSKFIENDKALRRRFNCVTINPFTSKETY 691
Query 123 SILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSHNK 182
+LKK+K++YE++H + +D L+ IV+++ Y +FPDKA+D+LDEA ++K+ + K
Sbjct 692 KLLKKIKYNYEKYHNIYYTDDSLKSIVSLTEDYLPTANFPDKAIDILDEAGVYQKIKYEK 751
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 150 bits (380), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 94/243 (38%), Positives = 141/243 (58%), Gaps = 14/243 (5%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP L+GRR+ISLD+ L AG RGEFE+R+K ++ +Q + DV++F+DEIHT+VG
Sbjct 373 GDVPDSLKGRRVISLDMAALIAGAKYRGEFEERLKAVLKEVQDAEGDVVMFIDEIHTVVG 432
Query 61 AGKTSGSSDAT--QILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSK 118
AG +LK LARGE +GATT EY+ +IEKD A RRFQ ++VE P
Sbjct 433 AGAGGEGGAMDAGNMLKPMLARGEFRCIGATTTNEYRQYIEKDKALERRFQKVLVEEPQV 492
Query 119 ERALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKV 178
+SIL+ +K YE HGV + + L N++ +Y R PDKA+DL+DEA + K+
Sbjct 493 SETISILRGLKDRYEVHHGVRILDSALVEAANLAHRYISDRFLPDKAIDLVDEAAARLKI 552
Query 179 SH----------NKKLVLLKMQIADMESNAADATAEDLQK--LKEELAALEALTADGRRL 226
+++L+ L+M+ ++ + + E+ +K L+ + +E L AD +L
Sbjct 553 QVSSKPIQLDEIDRRLLQLEMEKISIQGDGRERMLEEQEKWRLRSVESQIERLKADQGKL 612
Query 227 VLE 229
E
Sbjct 613 TEE 615
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 52/84 (61%), Gaps = 4/84 (4%)
Query 202 TAEDLQK-LKEELAALEALTADGRRLV---LELHDVAHILHLWTGIPLGKMTEDEVSRVL 257
T DL++ LKE + TA G+R++ + + D+A ++ +WTGIP+ ++ + E ++L
Sbjct 659 TLPDLERQLKEAEEQYKESTAGGKRMLRDEVTVDDIATVVAMWTGIPVTRLKQSEKEKLL 718
Query 258 RLADILSLRVIGQHQAVKAVADAL 281
L L RV+GQ AV+ VA+A+
Sbjct 719 NLEKDLHRRVVGQDHAVQVVAEAI 742
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 144 bits (363), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 84/218 (38%), Positives = 130/218 (59%), Gaps = 15/218 (6%)
Query 3 VPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAG 62
VP L+G ++ SLDL L+AG +G+FE+R K ++ ++ K ++LF+DEIH L+G G
Sbjct 234 VPTILQGAKLFSLDLAALTAGAKYKGDFEERFKGVLKEIEESKTLIVLFIDEIHMLMGNG 293
Query 63 KTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERAL 122
K DA ILK L+RG++ ++GATT EY+ +EKD AF RRFQ I V PS + +
Sbjct 294 K----DDAANILKPALSRGQLKVIGATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTV 349
Query 123 SILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSHNK 182
+IL+ ++ YE HGV + + L ++ +Y R PD ALDL+D +C+ V+ +
Sbjct 350 AILRGLQPKYEIHHGVRILDSALVTAAQLAKRYLPYRRLPDSALDLVDISCAGVAVARDS 409
Query 183 K----------LVLLKMQIADMESNA-ADATAEDLQKL 209
K L L++++I +E + AD+T +D KL
Sbjct 410 KPEELDSKERQLQLIQVEIKALERDEDADSTTKDRLKL 447
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 58/114 (50%), Gaps = 14/114 (12%)
Query 182 KKLVLLKMQIADMESNAADATAEDLQ-----KLKEELAALEALTADGRRL---------V 227
KKL L+ + D E ATA DL+ +K+++ LE A+ R V
Sbjct 481 KKLDELENKALDAERRYDTATAADLRYFAIPDIKKQIEKLEDQVAEEERRAGANSMIQNV 540
Query 228 LELHDVAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
++ ++ TGIP+ K++E E +++ + LS V+GQ A+KAV++A+
Sbjct 541 VDSDTISETAARLTGIPVKKLSESENEKLIHMERDLSSEVVGQMDAIKAVSNAV 594
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 135 bits (340), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 80/200 (40%), Positives = 117/200 (58%), Gaps = 9/200 (4%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ + I SLD+ +L AGT RG+FEKR K ++ L++ + ILF+DEIHT++G
Sbjct 234 GDVPEVMADCTIYSLDIGSLLAGTKYRGDFEKRFKALLKQLEQ-DTNSILFIDEIHTIIG 292
Query 61 AGKTSGSS-DATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKE 119
AG SG DA ++K L+ G+I ++G+TT +E+ EKD A RRFQ I + PS E
Sbjct 293 AGAASGGQVDAANLIKPLLSSGKIRVIGSTTYQEFSNIFEKDRALARRFQKIDITEPSIE 352
Query 120 RALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKK-- 177
+ I+ +K YE H V + V ++ +Y R PDKA+D++DEA + +
Sbjct 353 ETVQIINGLKPKYEAHHDVRYTAKAVRAAVELAVKYINDRHLPDKAIDVIDEAGARARLM 412
Query 178 -VSHNKKLVLLKMQIADMES 196
VS KK V +AD+ES
Sbjct 413 PVSKRKKTV----NVADIES 428
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 134 bits (338), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 76/184 (41%), Positives = 108/184 (58%), Gaps = 1/184 (0%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G V + + +R+ LD+ L AG RG+FE+R+ +I + + K++IL +DE H LVG
Sbjct 330 GHVLEKMLNKRLRQLDVGLLVAGARFRGQFEERLTRLIEEI-KNAKNIILVIDEAHMLVG 388
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA +LK LARGEI + TT +EY+ H EKDAA CRRFQ I V+ PS +
Sbjct 389 AGAGEGALDAANLLKPTLARGEIQCIAITTPKEYQKHFEKDAALCRRFQPIHVKEPSDKD 448
Query 121 ALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKKVSH 180
IL +FH V+ D + + S Q+ +R PDKA+D+LDEA S K+
Sbjct 449 TQIILNATAEACGRFHNVKYNMDAVAAALKYSKQFIPERYLPDKAIDILDEAGSLAKIRF 508
Query 181 NKKL 184
++L
Sbjct 509 YEQL 512
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 233 VAHILHLWTGIPLGKMTEDEVSRVLRLADILSLRVIGQHQAVKAVADAL 281
VA ++ WTG+PL K+T+ E+ + L L V+G +AVK +A A+
Sbjct 627 VAEVVSNWTGVPLKKLTQGEIEAIRNLEQELHKSVVGHEEAVKNIAKAI 675
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 103 bits (257), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 70/225 (31%), Positives = 116/225 (51%), Gaps = 9/225 (4%)
Query 12 IISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDAT 71
+ SLDL L +G RGE E ++K I ++ K ILF+DEIH L+ + T
Sbjct 233 VYSLDLCRLYSGQGTRGELEAKLKSIFDTVKNGKS--ILFIDEIHHLIQ--NQENGVNVT 288
Query 72 QILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKHH 131
+LK + + ++G+TT +EY + +D AF RRF+ + + S + L+IL +
Sbjct 289 NLLKPIMTSTLVKIIGSTTAKEYHQYFRRDRAFERRFEILRLHENSADETLAILHGSRPS 348
Query 132 YEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEAC---SWKKVSHNKKLVLLK 188
E +HGV++ +D L V +S ++ R PDKA+DLLDEA K+ N LL+
Sbjct 349 LEDYHGVKITDDALVASVELSTRFIPNRYLPDKAIDLLDEAAMLSKRKRSCSNDLFELLR 408
Query 189 MQIADMESNAADATAEDLQKLKEELAALEALTADGRRLVLELHDV 233
+ + + + D +L L+ +E++ + + L +L DV
Sbjct 409 HKTGIISALLRGKSGND--QLNRVLSEIESMESRYKSLHGKLRDV 451
> ath:AT3G45450 Clp amino terminal domain-containing protein
Length=341
Score = 70.5 bits (171), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 65/124 (52%), Gaps = 17/124 (13%)
Query 1 GAVPKGLRGRRIISLDLLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVG 60
G VP+ ++G+ +N+ AG E R + I + + D+ILF+DE+H L+G
Sbjct 203 GDVPETIKGK-------MNV-AGNCGWNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIG 254
Query 61 AGKTSGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKER 120
AG G+ DA ILK L R E+ +Y+ HIE D A RRFQ + V P+ E
Sbjct 255 AGAVEGAIDAANILKPALERCEL---------QYRKHIENDPALERRFQPVKVPEPTVEE 305
Query 121 ALSI 124
A+ I
Sbjct 306 AIQI 309
> cpv:cgd7_2620 ClpB ATpase (bacterial), signal peptide
Length=1263
Score = 44.7 bits (104), Expect = 4e-04, Method: Composition-based stats.
Identities = 41/173 (23%), Positives = 80/173 (46%), Gaps = 4/173 (2%)
Query 7 LRGRRIISLDLLNL--SAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKT 64
LRG RIIS+ L +L S + + E+ + + +I+F D + + +
Sbjct 533 LRGYRIISIHLESLLESCKNTKKSLTEQIKIKFDELMGAYDGKIIVFTDNLFS--SFETS 590
Query 65 SGSSDATQILKVPLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSI 124
+GS I+K + RG + ++ + E YK+ EK+ F I ++ + +
Sbjct 591 TGSKRLYDIMKHYIVRGTLKVIATLSNENYKILAEKEIEVKSIFYTIEMKELNGIVSEVF 650
Query 125 LKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKALDLLDEACSWKK 177
+ +++ E G+ + DV+ + V M +Y + PD A++L++ A S K
Sbjct 651 ISGLRYQLELSTGIFINNDVIRVSVLMCHKYIENCVLPDDAVELINFAISMAK 703
> sce:YNL218W MGS1; Mgs1p; K07478 putative ATPase
Length=587
Score = 38.5 bits (88), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 15/84 (17%)
Query 9 GRRIISLDLLNLSAGTS-MRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGS 67
G R ++ A T +RG FEK KE Q K+ +LF+DEIH
Sbjct 204 GSRYFMIETSATKANTQELRGIFEKSKKE----YQLTKRRTVLFIDEIHRF--------- 250
Query 68 SDATQILKVP-LARGEIVLVGATT 90
+ Q L +P + G+I+L+GATT
Sbjct 251 NKVQQDLLLPHVENGDIILIGATT 274
> ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=981
Score = 36.2 bits (82), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 51/110 (46%), Gaps = 17/110 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARG------ 81
G+ EK K + +F + VI+FVDEI +L+GA S +AT+ ++
Sbjct 769 GDAEKLTKALFSFATKLAP-VIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRS 827
Query 82 ----EIVLVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILK 126
I+++GAT + D A RR + I V+ P E L ILK
Sbjct 828 KDSQRILILGATNRP-----FDLDDAVIRRLPRRIYVDLPDAENRLKILK 872
> tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12196
vacuolar protein-sorting-associated protein 4
Length=502
Score = 35.8 bits (81), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 18/99 (18%)
Query 24 TSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL----- 78
+ +GE EK ++ + + R ++ I+F+DEI ++ GA ++ G SD+++ +K
Sbjct 269 SKWQGESEKLVRSLFA-MARERRPSIIFIDEIDSMCGA-RSEGDSDSSRRIKTEFLVQMQ 326
Query 79 -----ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIV 112
A G +VL GAT + + L D+A RRF+ V
Sbjct 327 GLQKDAPGVLVL-GATNV-PWAL----DSAIRRRFERRV 359
> ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=476
Score = 35.4 bits (80), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 37/138 (26%), Positives = 63/138 (45%), Gaps = 19/138 (13%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------- 78
GE EK ++ + R+ VI FVDEI +L+ K+ G ++++ LK
Sbjct 274 GEGEKLVRALFGVASCRQPAVI-FVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDS 332
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKHHYEQFHG 137
+I+L+GAT + E D A RR + + + PS E I++ + F
Sbjct 333 GSEQILLIGATNRPQ-----ELDEAARRRLTKRLYIPLPSSEARAWIIQNLLKKDGLFT- 386
Query 138 VELPEDVLELIVNMSAQY 155
L +D + +I N++ Y
Sbjct 387 --LSDDDMNIICNLTEGY 402
> dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog
b-like; K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 35.0 bits (79), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 16/110 (14%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------- 78
GE EK +K + T L R K I+F+DEI +L G+ ++ S+A + +K
Sbjct 203 GESEKLVKSLFT-LAREHKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGN 260
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKV 128
I+++GAT + + L D+A RRF+ + +E A S + K+
Sbjct 261 DNEGILVLGATNI-PWTL----DSAIRRRFEKRIYIPLPEEHARSFMFKL 305
> dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 16/110 (14%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------- 78
GE EK +K + T L R K I+F+DEI +L G+ ++ S+A + +K
Sbjct 203 GESEKLVKSLFT-LAREHKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGN 260
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKV 128
I+++GAT + + L D+A RRF+ + +E A S + K+
Sbjct 261 DNEGILVLGATNI-PWTL----DSAIRRRFEKRIYIPLPEEHARSFMFKL 305
> sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-ATPase
involved in multivesicular body (MVB) protein sorting,
ATP-bound Vps4p localizes to endosomes and catalyzes ESCRT-III
disassembly and membrane release; ATPase activity is activated
by Vta1p; regulates cellular sterol metabolism; K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 50/92 (54%), Gaps = 18/92 (19%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------- 78
GE EK +K++ + R K I+F+DE+ L G + G S+A++ +K L
Sbjct 208 GESEKLVKQLFA-MARENKPSIIFIDEVDALTGT-RGEGESEASRRIKTELLVQMNGVGN 265
Query 79 -ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 109
++G +VL GAT + ++L D+A RRF+
Sbjct 266 DSQGVLVL-GATNI-PWQL----DSAIRRRFE 291
> ath:AT5G52882 ATP binding / nucleoside-triphosphatase/ nucleotide
binding
Length=829
Score = 35.0 bits (79), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 38/123 (30%), Positives = 58/123 (47%), Gaps = 19/123 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR------- 80
GE EK ++ + T L + I+FVDE+ +++G G +A + +K
Sbjct 593 GEDEKNVRALFT-LAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMT 651
Query 81 --GEIVLVGATTLEEYKLHIEKDAAFCRRFQ-NIVVEAP---SKERAL-SILKKVKHHYE 133
GE +LV A T + L D A RRF+ I+V P S+E+ L ++L K K
Sbjct 652 KPGERILVLAATNRPFDL----DEAIIRRFERRIMVGLPSIESREKILRTLLSKEKTENL 707
Query 134 QFH 136
FH
Sbjct 708 DFH 710
> ath:AT1G64110 AAA-type ATPase family protein
Length=827
Score = 34.7 bits (78), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 51/109 (46%), Gaps = 15/109 (13%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR------- 80
GE EK ++ + T L + I+FVDE+ +++G G +A + +K
Sbjct 595 GEDEKNVRALFT-LASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMT 653
Query 81 --GEIVLVGATTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILK 126
GE +LV A T + L D A RRF+ I+V P+ E IL+
Sbjct 654 KPGERILVLAATNRPFDL----DEAIIRRFERRIMVGLPAVENREKILR 698
> ath:AT4G28000 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=830
Score = 33.5 bits (75), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 51/109 (46%), Gaps = 15/109 (13%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLAR------- 80
GE EK ++ + T L + I+FVDE+ +++G G +A + +K
Sbjct 594 GEDEKNVRALFT-LAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMS 652
Query 81 --GEIVLVGATTLEEYKLHIEKDAAFCRRFQ-NIVVEAPSKERALSILK 126
G+ +LV A T + L D A RRF+ I+V PS E IL+
Sbjct 653 NAGDRILVLAATNRPFDL----DEAIIRRFERRIMVGLPSVESREKILR 697
> sce:YLL034C RIX7; Putative ATPase of the AAA family, required
for export of pre-ribosomal large subunits from the nucleus;
distributed between the nucleolus, nucleoplasm, and nuclear
periphery depending on growth conditions; K14571 ribosome
biogenesis ATPase
Length=837
Score = 33.5 bits (75), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 56/127 (44%), Gaps = 20/127 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLA-------R 80
GE E+ ++++ T R ++F DE+ LV TS S +++++ L R
Sbjct 609 GESERSIRQVFT-RARASVPCVIFFDELDALVPRRDTSLSESSSRVVNTLLTELDGLNDR 667
Query 81 GEIVLVGATTLEEYKLHIEKDAAFCRRF---QNIVVEAPSKERALSILKKVKHHYEQFHG 137
I ++GAT + D A R +++ +E P+ E L I+K + HG
Sbjct 668 RGIFVIGATNRPDM-----IDPAMLRPGRLDKSLFIELPNTEEKLDIIKTLTKS----HG 718
Query 138 VELPEDV 144
L DV
Sbjct 719 TPLSSDV 725
> ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1001
Score = 33.1 bits (74), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 17/110 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARG------ 81
G+ EK K + +F + VI+FVDE+ +L+GA + +AT+ ++
Sbjct 789 GDAEKLTKALFSFASKLAP-VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRS 847
Query 82 ----EIVLVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILK 126
I+++GAT + D A RR + I V+ P E L ILK
Sbjct 848 KDSQRILILGATNRP-----FDLDDAVIRRLPRRIYVDLPDAENRLKILK 892
> mmu:67299 Dock7, 3110056M06Rik, Gm430, m, mKIAA1771; dedicator
of cytokinesis 7
Length=2098
Score = 33.1 bits (74), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query 118 KERALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKA 165
KE A++ L ++ H E F+G EDVLE+I + S DK + P+KA
Sbjct 1821 KEPAITKLAEISHRLEGFYGERFGEDVLEVIKD-SNPVDKCKLDPNKA 1867
> cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting
factor family member (vps-4); K12196 vacuolar protein-sorting-associated
protein 4
Length=430
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 14/90 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDA--------TQILKVPLA 79
GE EK +K + L R K I+F+DEI +L A + S A Q+ V L
Sbjct 194 GESEKLVKNLFA-LAREHKPSIIFIDEIDSLCSARSDNESESARRIKTEFMVQMQGVGLN 252
Query 80 RGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 109
I+++GAT + + L D+A RRF+
Sbjct 253 NDGILVLGATNI-PWIL----DSAIRRRFE 277
> hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting
4 homolog B (S. cerevisiae); K12196 vacuolar protein-sorting-associated
protein 4
Length=444
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 48/91 (52%), Gaps = 16/91 (17%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPL 78
GE EK +K + L R K I+F+DEI +L G+ ++ S+A + +K V +
Sbjct 210 GESEKLVKNLFQ-LARENKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGV 267
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 109
I+++GAT + + L D+A RRF+
Sbjct 268 DNDGILVLGATNI-PWVL----DSAIRRRFE 293
> tgo:TGME49_028070 hypothetical protein
Length=3891
Score = 32.7 bits (73), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 123 SILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQR 159
S+++ +KHHYE+ H ELP D ++ + QR
Sbjct 1761 SLIRALKHHYEKIHKTELPADSDTFVLPWEKEKRLQR 1797
> mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=444
Score = 32.7 bits (73), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 48/91 (52%), Gaps = 16/91 (17%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPL 78
GE EK +K + L R K I+F+DEI +L G+ ++ S+A + +K V +
Sbjct 210 GESEKLVKNLFQ-LARENKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGV 267
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 109
I+++GAT + + L D+A RRF+
Sbjct 268 DNDGILVLGATNI-PWVL----DSAIRRRFE 293
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 32.3 bits (72), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 53/128 (41%), Gaps = 22/128 (17%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPL--------A 79
GE EK + + + R I+FVDEI +L G S+A++ +K L
Sbjct 207 GESEKLVSNLFE-MARESAPSIIFVDEIDSLCGTRGEGNESEASRRIKTELLVQMQGVGH 265
Query 80 RGEIVLVGATTLEEYKLHIEKDAAFCRRF-QNIVVEAPSKERALSILKKVKHHYEQFHGV 138
E VLV A T Y L D A RRF + I + P K + H + H
Sbjct 266 NDEKVLVLAATNTPYAL----DQAIRRRFDKRIYIPLPE--------AKARQHMFKVHLG 313
Query 139 ELPEDVLE 146
+ P ++ E
Sbjct 314 DTPHNLTE 321
> dre:405856 MGC85976; zgc:85976; K07478 putative ATPase
Length=546
Score = 32.3 bits (72), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 55/111 (49%), Gaps = 11/111 (9%)
Query 17 LLNLSAGTSMRGEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKV 76
+ LSA ++ + + +K+ L+ K+ +LF+DEIH + + S T + V
Sbjct 183 FVTLSATSASVSDVREVIKQAQNELRLCKRKTVLFIDEIH------RFNKSQQDTFLPHV 236
Query 77 PLARGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKK 127
G I L+GATT E + ++A R + +V+E S E SIL++
Sbjct 237 EC--GTITLIGATT-ENPSFQV--NSALLSRCRVLVLERLSVEAVGSILRR 282
> dre:567576 spectrin repeat containing, nuclear envelope 1-like
Length=8621
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGEIVLVG 87
G+F ++ +E++T+ Q KK + +D+ H ++ K+ +S + L L I +
Sbjct 798 GDFTQKCQELVTYQQSCKK-CLSVIDKNHQVIL--KSLDTSKNLKHLDTSLLERRITELQ 854
Query 88 ATT------LEEYKLHIEKDAAFCRRFQNIVVE 114
A++ E+K H+E +++ +RF+ VE
Sbjct 855 ASSQGMVKETTEWKQHVEANSSLMKRFEESRVE 887
> xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog
B; K12196 vacuolar protein-sorting-associated protein 4
Length=442
Score = 32.0 bits (71), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 55/109 (50%), Gaps = 17/109 (15%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPL 78
GE EK +K + L R K I+F+DEI +L G+ ++ S+A + +K V +
Sbjct 208 GESEKLVKNLFQ-LAREHKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGV 265
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQN-IVVEAPSKERALSILK 126
I+++GAT + + L D+A RRF+ I + P + ++ K
Sbjct 266 DNEGILVLGATNI-PWVL----DSAIRRRFEKRIYIPLPEEHARAAMFK 309
> dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sorting
4a (yeast); K12196 vacuolar protein-sorting-associated
protein 4
Length=440
Score = 32.0 bits (71), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 51/106 (48%), Gaps = 14/106 (13%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDA--------TQILKVPLA 79
GE EK +K + L R+ K I+F+DE+ +L G+ + S A Q+ V
Sbjct 202 GESEKLVKNLFD-LARQHKPSIIFIDEVDSLCGSRNENESEAARRIKTEFLVQMQGVGNN 260
Query 80 RGEIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSIL 125
I+++GAT + + L DAA RRF+ + +E A S +
Sbjct 261 NDGILVLGATNI-PWVL----DAAIRRRFEKRIYIPLPEEPARSAM 301
> hsa:85440 DOCK7, KIAA1771, ZIR2; dedicator of cytokinesis 7
Length=2109
Score = 32.0 bits (71), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query 118 KERALSILKKVKHHYEQFHGVELPEDVLELIVNMSAQYDKQRSFPDKA 165
KE A++ L ++ H E F+G EDV+E+I + S DK + P+KA
Sbjct 1832 KEPAITKLAEISHRLEGFYGERFGEDVVEVIKD-SNPVDKCKLDPNKA 1878
> xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-associated
protein 4
Length=443
Score = 31.6 bits (70), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 48/91 (52%), Gaps = 16/91 (17%)
Query 28 GEFEKRMKEIITFLQRRKKDVILFVDEIHTLVGAGKTSGSSDATQILK---------VPL 78
GE EK +K + L R K I+F+DEI +L G+ ++ S+A + +K V +
Sbjct 209 GESEKLVKNLFQ-LAREHKPSIIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGV 266
Query 79 ARGEIVLVGATTLEEYKLHIEKDAAFCRRFQ 109
I+++GAT + + L D+A RRF+
Sbjct 267 DNEGILVLGATNI-PWVL----DSAIRRRFE 292
Lambda K H
0.319 0.135 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10975721104
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40