bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0008_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
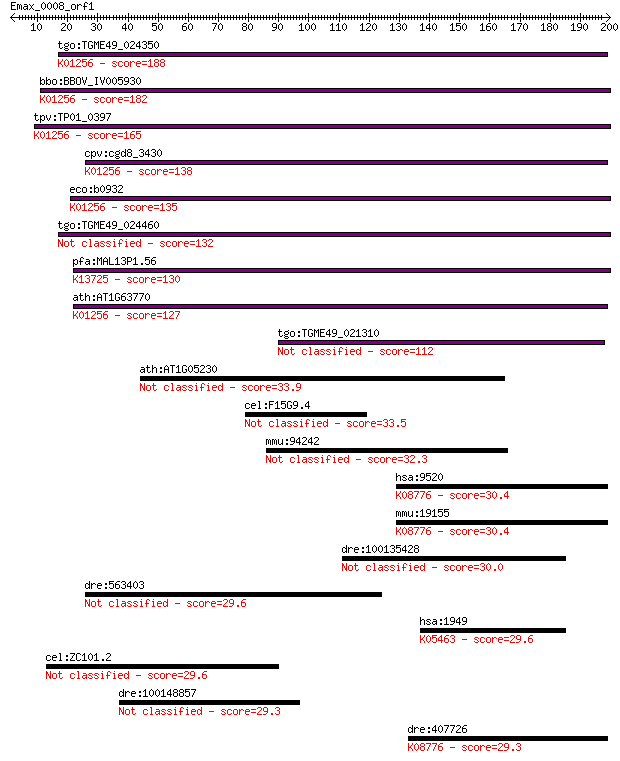
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 188 9e-48
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 182 7e-46
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 165 8e-41
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 138 1e-32
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 135 8e-32
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 132 1e-30
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 130 4e-30
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 127 4e-29
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 112 6e-25
ath:AT1G05230 HDG2; HDG2 (HOMEODOMAIN GLABROUS 2); DNA binding... 33.9 0.38
cel:F15G9.4 him-4; High Incidence of Males (increased X chromo... 33.5 0.51
mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,... 32.3 1.2
hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin s... 30.4 4.0
mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku; ... 30.4 4.7
dre:100135428 lrrc9; leucine rich repeat containing 9 30.0
dre:563403 hypothetical LOC563403 29.6 6.6
hsa:1949 EFNB3, EFL6, EPLG8, LERK8; ephrin-B3; K05463 ephrin-B 29.6 7.3
cel:ZC101.2 unc-52; UNCoordinated family member (unc-52) 29.6 8.0
dre:100148857 similar to Retinoid- and fatty-acid binding prot... 29.3 8.8
dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminope... 29.3 9.3
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 188 bits (478), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 96/182 (52%), Positives = 126/182 (69%), Gaps = 0/182 (0%)
Query 17 KQRGRPGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEE 76
K +P EK R +YKP+DF +D VDLDF L + TKV +T+ M R T PTDLVL+GE+
Sbjct 515 KPTKQPVEKHRLDYKPTDFLIDFVDLDFDLYDDRTKVTSTLTMHRREQTPPTDLVLDGED 574
Query 77 LDLVSLTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTLQTQVDI 136
L+L S+ L+G +S A + Y L DG L+I+ +LP+EA + F ++T V +
Sbjct 575 LELESVELDGNALSMHSTETQKAGDKRVYSLDVDGRLVIAADLLPQEAEKKFKVKTVVYV 634
Query 137 HPKANLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSN 196
PK NL+L GLY SG+ LVTQCEAEGFRRITYFLDRPDV++ + VRL A+++ PVLLSN
Sbjct 635 RPKENLQLMGLYKSGALLVTQCEAEGFRRITYFLDRPDVMSLFKVRLAADEKACPVLLSN 694
Query 197 GN 198
GN
Sbjct 695 GN 696
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 182 bits (462), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 98/189 (51%), Positives = 128/189 (67%), Gaps = 11/189 (5%)
Query 11 LTAIRAKQRGRPGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDL 70
LT AKQ E FR +Y P+ + +DSV LDF L ET T V A ++M R GT+P DL
Sbjct 22 LTLKPAKQHV---EIFRKDYTPTIYDIDSVFLDFDLHETATVVKAELLMHRKEGTAPADL 78
Query 71 VLNGEELDLVSLTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTL 130
VL+G+ELD S++++GK + + P + GY + DG L I + LP +AGESF +
Sbjct 79 VLHGDELDCRSVSVDGKPLEN--RPLS------GYHIDDDGFLNIPVSFLPSKAGESFRV 130
Query 131 QTQVDIHPKANLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKY 190
T+V I+P ANL+L GLY + TQCE+ GFRRITYFLDRPDVL++Y VRL A+K++Y
Sbjct 131 NTEVVINPTANLQLSGLYKNSQLFTTQCESHGFRRITYFLDRPDVLSRYRVRLRADKDQY 190
Query 191 PVLLSNGNK 199
PVLLSNGNK
Sbjct 191 PVLLSNGNK 199
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 165 bits (418), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 85/191 (44%), Positives = 125/191 (65%), Gaps = 8/191 (4%)
Query 9 SELTAIRAKQRGRPGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPT 68
SE +++ K + E FR +YKP +F V++V L+F LE+T TKV + ++M+R T P
Sbjct 139 SETSSVTLKPVKKFVEIFRKDYKPPEFDVENVYLEFDLEDTETKVRSKLLMRRRPNTLPG 198
Query 69 DLVLNGEELDLVSLTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESF 128
+L LNG++L + ++G ++ + GY L DG++ + A LPK+ +F
Sbjct 199 NLTLNGDDLTCNFVAVDGVELKNVP--------VSGYTLDVDGNMTVPSAFLPKDDSRTF 250
Query 129 TLQTQVDIHPKANLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKE 188
T++T V I+PK N KL GLY S +A TQCE GFRRITY+LDRPDVL+ + VR++A+K+
Sbjct 251 TVETHVTINPKNNTKLVGLYKSSAAFCTQCEPHGFRRITYYLDRPDVLSSFRVRVQADKK 310
Query 189 KYPVLLSNGNK 199
YPVLLSNGN+
Sbjct 311 LYPVLLSNGNR 321
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 82/176 (46%), Positives = 110/176 (62%), Gaps = 12/176 (6%)
Query 26 FRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPT-DLVLNGEELDLVSLTL 84
+R EYK +F +D V+LD +++ T V + IIM+R +S DL L+G+ L LVS+ L
Sbjct 31 YRKEYKVPNFLIDHVNLDINIKDDVTVVSSVIIMRRNPNSSFRGDLSLDGDCLKLVSVKL 90
Query 85 NGKKISDSGNPNAMADGEEGYRL--GSDGSLIISKAVLPKEAGESFTLQTQVDIHPKANL 142
NG + S +GY G DG L+IS +LP + + FTL+T V+I P N
Sbjct 91 NGVILEKSL--------YKGYFQPDGPDGKLVISSNLLPNK-DQQFTLETVVEIFPDRNT 141
Query 143 KLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSNGN 198
K GLY S TQCE++GFRRIT+FLDRPDV+ K+ VR+E +K K PVLLSNGN
Sbjct 142 KNMGLYYSAGVYSTQCESDGFRRITFFLDRPDVMCKFRVRIEGDKTKSPVLLSNGN 197
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 135 bits (341), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 82/179 (45%), Positives = 107/179 (59%), Gaps = 21/179 (11%)
Query 21 RPGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLV 80
+P K+R +Y+ D+ + +DL F L+ T V A R G S L LNGE+L LV
Sbjct 4 QPQAKYRHDYRAPDYQITDIDLTFDLDAQKTVVTAVSQAVRH-GASDAPLRLNGEDLKLV 62
Query 81 SLTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPKA 140
S+ +N + + A EE +G+L+IS LP E FTL+ +I P A
Sbjct 63 SVHINDEPWT--------AWKEE------EGALVISN--LP----ERFTLKIINEISPAA 102
Query 141 NLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSNGNK 199
N L GLY SG AL TQCEAEGFR ITY+LDRPDVLA++T ++ A+K KYP LLSNGN+
Sbjct 103 NTALEGLYQSGDALCTQCEAEGFRHITYYLDRPDVLARFTTKIIADKIKYPFLLSNGNR 161
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 132 bits (331), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 78/198 (39%), Positives = 117/198 (59%), Gaps = 21/198 (10%)
Query 17 KQRGRPGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEE 76
+++ P R +YKP +F ++ + LDF L++ +T V A + + R GT P +LVL+G+E
Sbjct 49 EEKEEPKAVVRLDYKPPNFYLEDIVLDFNLDDEDTTVEALLTLYRRAGTEPMNLVLDGDE 108
Query 77 LDLVSLTLNGKKISDSGNPNAM---------------ADGEEGYRLGSDGSLIISKAVLP 121
S+ L ++ + +P+ D + +R+ DG+L++SK +LP
Sbjct 109 ----SVILKSISLATTSSPSLRGQGRVTFTELDGQMPTDSQFQFRI-VDGNLMVSKFLLP 163
Query 122 KEAGESFTLQTQVDIHPKANLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTV 181
+EA F ++T V I+PKANL L GLY +G VT E GFRRITY +DRPDVLA YTV
Sbjct 164 REAEVRFYIRTLVSINPKANLALFGLYKAGDIFVTLNEPSGFRRITYGVDRPDVLATYTV 223
Query 182 RLEANKEKYPVLLSNGNK 199
+ A + P+LLSNG+K
Sbjct 224 TVTAPRH-LPILLSNGDK 240
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 130 bits (326), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 74/178 (41%), Positives = 107/178 (60%), Gaps = 14/178 (7%)
Query 22 PGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLVS 81
P +R +YKPS F +++V L+ + + T V + + M + DLV +G L +
Sbjct 196 PKIHYRKDYKPSGFIINNVTLNINIHDNETIVRSVLDMDISKHNVGEDLVFDGVGLKINE 255
Query 82 LTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPKAN 141
+++N KK + +GEE Y ++ I SK V PK F ++V IHP+ N
Sbjct 256 ISINNKK---------LVEGEE-YTYDNEFLTIFSKFV-PKS---KFAFSSEVIIHPETN 301
Query 142 LKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSNGNK 199
L GLY S + +V+QCEA GFRRIT+F+DRPD++AKY V + A+KEKYPVLLSNG+K
Sbjct 302 YALTGLYKSKNIIVSQCEATGFRRITFFIDRPDMMAKYDVTVTADKEKYPVLLSNGDK 359
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 127 bits (318), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 75/177 (42%), Positives = 99/177 (55%), Gaps = 16/177 (9%)
Query 22 PGEKFRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLVS 81
P E F Y D+ ++VDL F L E T V + I + + S LVL+G +L L+S
Sbjct 96 PKEIFLKNYTKPDYYFETVDLSFSLGEEKTIVSSKIKVSPRVKGSSAALVLDGHDLKLLS 155
Query 82 LTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPKAN 141
+ + GK + E Y+L S + S LP E ESF L+ +I+P N
Sbjct 156 VKVEGKLLK-----------EGDYQLDSRHLTLPS---LPAE--ESFVLEIDTEIYPHKN 199
Query 142 LKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSNGN 198
L GLY S TQCEAEGFR+IT++ DRPD++AKYT R+E +K YPVLLSNGN
Sbjct 200 TSLEGLYKSSGNFCTQCEAEGFRKITFYQDRPDIMAKYTCRVEGDKTLYPVLLSNGN 256
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 112 bits (281), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 55/108 (50%), Positives = 71/108 (65%), Gaps = 0/108 (0%)
Query 90 SDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPKANLKLRGLYA 149
S S + M G + GSL+I K +LP E+ + F ++TQV I P+ N +L GLY
Sbjct 99 SSSPSDKEMISPRPGIAVEKSGSLLICKDILPSESEQRFVVKTQVRISPQDNSRLSGLYV 158
Query 150 SGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLEANKEKYPVLLSNG 197
S LVT EA+GFRRIT+FLDRPDV+ ++ VRL A K+ YPVLLSNG
Sbjct 159 SDGVLVTHNEAQGFRRITFFLDRPDVMTQWRVRLTARKQDYPVLLSNG 206
> ath:AT1G05230 HDG2; HDG2 (HOMEODOMAIN GLABROUS 2); DNA binding
/ transcription factor
Length=719
Score = 33.9 bits (76), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 53/121 (43%), Gaps = 20/121 (16%)
Query 44 FFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLVSLTLNGKKISDSGNPNAMADGEE 103
L+E+ T A+ ++ + ++VLNG + D V+L +G I GN N+ A G +
Sbjct 611 LILQESCTDPTASFVIYAPVDIVAMNIVLNGGDPDYVALLPSGFAILPDGNANSGAPGGD 670
Query 104 GYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPKANLKLRGLYASGSALVTQCEAEGF 163
G GSL+ + Q VD P A L L G A+ + L+ C E
Sbjct 671 G------GSLL------------TVAFQILVDSVPTAKLSL-GSVATVNNLIA-CTVERI 710
Query 164 R 164
+
Sbjct 711 K 711
> cel:F15G9.4 him-4; High Incidence of Males (increased X chromosome
loss) family member (him-4)
Length=5213
Score = 33.5 bits (75), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 24/40 (60%), Gaps = 9/40 (22%)
Query 79 LVSLTLNGKKISDSGNPNAMADGEEGYRLGSDGSLIISKA 118
++S TLNG N + DGE G+ +G+DG+L I KA
Sbjct 4205 IISWTLNG---------NDIKDGENGHTIGADGTLHIEKA 4235
> mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,
Tinagl; tubulointerstitial nephritis antigen-like 1
Length=466
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 14/86 (16%)
Query 86 GKKISDSGNPNAMADGEE------GYRLGSDGSLIISKAVLPKEAGESFTLQTQVDIHPK 139
GK+ + S PN D + YRLGSD I+ KE E+ +Q +++H
Sbjct 318 GKRQATSRCPNGQVDSNDIYQVTPAYRLGSDEKEIM------KELMENGPVQALMEVHED 371
Query 140 ANLKLRGLYASGSALVTQCEAEGFRR 165
L RG+Y+ V+Q E +RR
Sbjct 372 FFLYQRGIYS--HTPVSQGRPEQYRR 395
> hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin
sensitive (EC:3.4.11.14); K08776 puromycin-sensitive aminopeptidase
[EC:3.4.11.-]
Length=919
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 38/84 (45%), Gaps = 16/84 (19%)
Query 129 TLQT-----QVDIHPKANLKLRGLYASGS---------ALVTQCEAEGFRRITYFLDRPD 174
TLQT ++D + N K++G Y S A VTQ EA RR D P
Sbjct 136 TLQTGTGTLKIDFVGELNDKMKGFYRSKYTTPSGEVRYAAVTQFEATDARRAFPCWDEPA 195
Query 175 VLAKYTVRLEANKEKYPVLLSNGN 198
+ A + + L K++ V LSN N
Sbjct 196 IKATFDISLVVPKDR--VALSNMN 217
> mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku;
aminopeptidase puromycin sensitive (EC:3.4.11.14); K08776
puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=920
Score = 30.4 bits (67), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 38/84 (45%), Gaps = 16/84 (19%)
Query 129 TLQT-----QVDIHPKANLKLRGLYASGS---------ALVTQCEAEGFRRITYFLDRPD 174
TLQT ++D + N K++G Y S A VTQ EA RR D P
Sbjct 137 TLQTGTGTLKIDFVGELNDKMKGFYRSRYTTPAGEVRYAAVTQFEATDARRAFPCWDEPA 196
Query 175 VLAKYTVRLEANKEKYPVLLSNGN 198
+ A + + L K++ V LSN N
Sbjct 197 IKATFDISLVVPKDR--VALSNMN 218
> dre:100135428 lrrc9; leucine rich repeat containing 9
Length=1440
Score = 30.0 bits (66), Expect = 4.8, Method: Composition-based stats.
Identities = 26/92 (28%), Positives = 35/92 (38%), Gaps = 22/92 (23%)
Query 111 GSLIISKAVLPKEAGESFTLQTQVDIHPKANLKLRGLYAS------------------GS 152
G LIISK L + L D +PKAN +Y S S
Sbjct 495 GQLIISKVFLGRSVAVKDGLPIDCDHYPKAN----SVYLSTSSKQQSHTAQTTPDMLCSS 550
Query 153 ALVTQCEAEGFRRITYFLDRPDVLAKYTVRLE 184
+L C+ + +R+ Y D VL +Y V E
Sbjct 551 SLPNSCDCKQQQRLWYMFDHEMVLPEYLVDFE 582
> dre:563403 hypothetical LOC563403
Length=913
Score = 29.6 bits (65), Expect = 6.6, Method: Composition-based stats.
Identities = 29/98 (29%), Positives = 38/98 (38%), Gaps = 7/98 (7%)
Query 26 FRDEYKPSDFAVDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLVSLTLN 85
F DE S FA D V EE +T TD G+E DL L
Sbjct 307 FPDELDTSFFAKDGVTH----EELSTYADEVFESPSEAAIKETDASRAGDERDLTGSALE 362
Query 86 GKKISDSGNPNAMADGEEGYRLGSDGSLIISKAVLPKE 123
++ S + M E G+R DG+L+ K L +E
Sbjct 363 KTELERS---HLMLPLERGWRKAKDGALVQPKVRLRQE 397
> hsa:1949 EFNB3, EFL6, EPLG8, LERK8; ephrin-B3; K05463 ephrin-B
Length=340
Score = 29.6 bits (65), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 137 HPKANLKLRGLYASGSALVTQCEAEGFRRITYFLDRPDVLAKYTVRLE 184
H N + LY G A +CEA + DRPD+ ++T++ +
Sbjct 71 HSSPNYEFYKLYLVGGAQGRRCEAPPAPNLLLTCDRPDLDLRFTIKFQ 118
> cel:ZC101.2 unc-52; UNCoordinated family member (unc-52)
Length=3375
Score = 29.6 bits (65), Expect = 8.0, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 43/85 (50%), Gaps = 9/85 (10%)
Query 13 AIRAKQRGRPGEKFRDEYKPSD------FAVDSVDLDFFLEETN--TKVIATIIMQRTIG 64
++R +++GR G+ D Y+ D A+ +VD + F+ +K + +G
Sbjct 3273 SVRFERKGREGQMRIDNYREVDGRSTGILAMLNVDGNIFVGGVPDISKATGGLFSNNFVG 3332
Query 65 TSPTDLVLNGEELDLVSLTLNGKKI 89
D+ LNG +LDL++ ++GK +
Sbjct 3333 CI-ADVELNGVKLDLMATAIDGKNV 3356
> dre:100148857 similar to Retinoid- and fatty-acid binding protein
CG11064-PA
Length=689
Score = 29.3 bits (64), Expect = 8.8, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 26/60 (43%), Gaps = 0/60 (0%)
Query 37 VDSVDLDFFLEETNTKVIATIIMQRTIGTSPTDLVLNGEELDLVSLTLNGKKISDSGNPN 96
V V L LE T TK ++++ + RT+ P DL N L + G+ P+
Sbjct 109 VSLVPLPGLLEVTETKTVSSLTLLRTLDVIPMDLAQNTGYLSSIVFEAEGQSSGRHAGPS 168
> dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminopeptidase
puromycin sensitive (EC:3.4.11.-); K08776 puromycin-sensitive
aminopeptidase [EC:3.4.11.-]
Length=872
Score = 29.3 bits (64), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 36/74 (48%), Gaps = 10/74 (13%)
Query 133 QVDIHPKANLKLRGLY-----ASGS---ALVTQCEAEGFRRITYFLDRPDVLAKYTVRLE 184
++D + N K++G Y +SG A VTQ EA RR D P + A + + L
Sbjct 101 KIDFVGELNDKMKGFYRSKYSSSGEVRYAAVTQFEATDARRAFPCWDEPAIKATFDITLI 160
Query 185 ANKEKYPVLLSNGN 198
K++ V LSN N
Sbjct 161 VPKDR--VALSNMN 172
Lambda K H
0.313 0.134 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5931269072
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40