bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3332_orf1
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
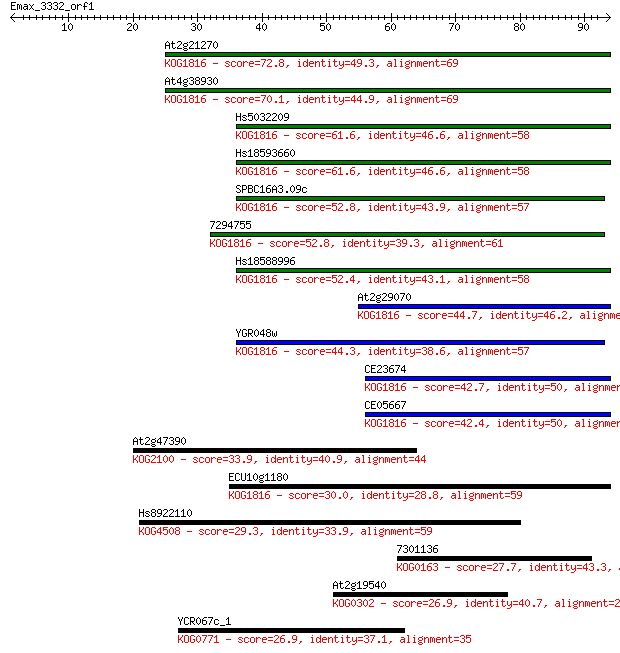
At2g21270 72.8 2e-13
At4g38930 70.1 9e-13
Hs5032209 61.6 3e-10
Hs18593660 61.6 3e-10
SPBC16A3.09c 52.8 1e-07
7294755 52.8 2e-07
Hs18588996 52.4 2e-07
At2g29070 44.7 4e-05
YGR048w 44.3 6e-05
CE23674 42.7 2e-04
CE05667 42.4 2e-04
At2g47390 33.9 0.076
ECU10g1180 30.0 1.1
Hs8922110 29.3 1.9
7301136 27.7 4.9
At2g19540 26.9 8.5
YCR067c_1 26.9 9.8
> At2g21270
Length=320
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 34/69 (49%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 25 FWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICN 84
F+ G G F SY YP SF K LESG+KI++PPSAL LA LHI +PM F + N
Sbjct 3 FFDGYHYHGTTFEQSYRCYPASFIDKPQLESGDKIIMPPSALDRLASLHIDYPMLFELRN 62
Query 85 PAKDKLTHC 93
+++THC
Sbjct 63 AGIERVTHC 71
> At4g38930
Length=315
Score = 70.1 bits (170), Expect = 9e-13, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 25 FWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICN 84
F+ G G F +Y YP SF K +ESG+KI++PPSAL LA L I +PM F + N
Sbjct 2 FYDGYAYHGTTFEQTYRCYPSSFIDKPQIESGDKIIMPPSALDRLASLQIDYPMLFELRN 61
Query 85 PAKDKLTHC 93
+ D +HC
Sbjct 62 ASTDSFSHC 70
> Hs5032209
Length=343
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y + VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCFSVSMLAWPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> Hs18593660
Length=307
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y + VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> SPBC16A3.09c
Length=342
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 36/60 (60%), Gaps = 3/60 (5%)
Query 36 FSASYTAYPVSF---AAKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLTH 92
F Y YPV+ + ++ G K++LPPSAL L+RL++S+PM F N A +K TH
Sbjct 32 FDTRYRCYPVAMIPGEERPNVNYGGKVILPPSALEKLSRLNVSYPMLFDFENEAAEKKTH 91
> 7294755
Length=316
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 38/64 (59%), Gaps = 3/64 (4%)
Query 32 DGGGFSASYTAYPVSFAA---KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKD 88
+G F A+Y + VS + +E G KI++PPSAL L RL++ +PM F++ N K
Sbjct 13 EGRNFHANYKCFSVSMLPGNERTDVEKGGKIIMPPSALDTLTRLNVEYPMLFKLTNVKKS 72
Query 89 KLTH 92
+ +H
Sbjct 73 RSSH 76
> Hs18588996
Length=288
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 39/62 (62%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y + VS A K +E G KI++ PSAL L++L+I++PM F++ + D++T
Sbjct 19 FSTHYRCFSVSTLAGPNDKSDVEKGGKIIMLPSALDQLSQLNITYPMLFKLTSKNLDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> At2g29070
Length=292
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 55 SGNKILLPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
S + ++PPSAL LA LHI +PM F++ N + +K +HC
Sbjct 7 SFEQFIMPPSALDRLASLHIEYPMLFQLSNVSVEKTSHC 45
> YGR048w
Length=361
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 4/61 (6%)
Query 36 FSASYTAYPVSFA----AKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
F + YP++ KD G KI LPPSAL L+ L+I +PM F++ ++T
Sbjct 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80
Query 92 H 92
H
Sbjct 81 H 81
> CE23674
Length=336
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 56 GNKILLPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
G KILLP SAL+ L + +I PM F++ N A ++THC
Sbjct 41 GGKILLPSSALNLLMQYNIPMPMLFKLTNMAVQRVTHC 78
> CE05667
Length=342
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 56 GNKILLPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
G KILLP SAL+ L + +I PM F++ N A ++THC
Sbjct 47 GGKILLPSSALNLLMQYNIPMPMLFKLTNMAVQRVTHC 84
> At2g47390
Length=955
Score = 33.9 bits (76), Expect = 0.076, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 20 RSMASFWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPP 63
RS+AS GG EDGGG S + + D L G LPP
Sbjct 71 RSLASACSGGAEDGGGTSNGSLSASATATEDDELAIGTGYRLPP 114
> ECU10g1180
Length=227
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 24/59 (40%), Gaps = 0/59 (0%)
Query 35 GFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
G S++ P F + G K+++P S L L I P F I + THC
Sbjct 10 GEKPSWSLRPTKFDGCNQNNFGGKVIVPQSVLVDLVSFQIQPPFTFEISHSDGIYRTHC 68
> Hs8922110
Length=510
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 21 SMASFWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKIL-------LPPSALHALARLH 73
S+ S GGG G S ++ Y F+ DH G+ ++ L PSA L L+
Sbjct 73 SIISEEAGGGSTFEGLSTAFHHY---FSKADHFRLGSVLVMLLQQPDLLPSAAQRLTALY 129
Query 74 ISWPMH 79
+ W M+
Sbjct 130 LLWEMY 135
> 7301136
Length=1253
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Query 61 LP-PSALHALARLHISWPMHFRICNPAKDKL 90
LP PS H A +H SW H+R+ P +L
Sbjct 535 LPKPSYSHFTAEVHKSWANHYRLGLPRSSRL 565
> At2g19540
Length=469
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 51 DHLESGNKILLPPSALHALARLHISWP 77
D LE G ++ PSA ++L H+ WP
Sbjct 37 DTLEDGEELQCDPSAYNSLHGFHVGWP 63
> YCR067c_1
Length=422
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 27 GGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILL 61
GGGG+ F TA V+F K H+ +I L
Sbjct 29 GGGGQFNSSFPNKITALKVNFQKKKHIRRFREITL 63
Lambda K H
0.325 0.141 0.481
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167934574
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40