bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3258_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
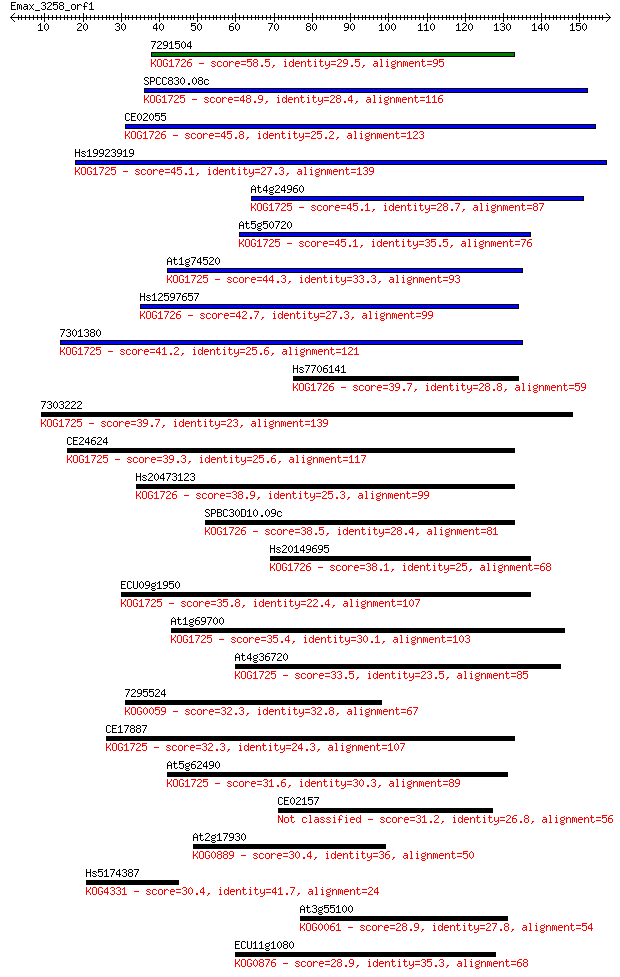
7291504 58.5 5e-09
SPCC830.08c 48.9 4e-06
CE02055 45.8 3e-05
Hs19923919 45.1 6e-05
At4g24960 45.1 6e-05
At5g50720 45.1 6e-05
At1g74520 44.3 9e-05
Hs12597657 42.7 3e-04
7301380 41.2 9e-04
Hs7706141 39.7 0.002
7303222 39.7 0.003
CE24624 39.3 0.003
Hs20473123 38.9 0.004
SPBC30D10.09c 38.5 0.005
Hs20149695 38.1 0.008
ECU09g1950 35.8 0.032
At1g69700 35.4 0.052
At4g36720 33.5 0.17
7295524 32.3 0.38
CE17887 32.3 0.43
At5g62490 31.6 0.74
CE02157 31.2 0.78
At2g17930 30.4 1.4
Hs5174387 30.4 1.5
At3g55100 28.9 4.4
ECU11g1080 28.9 4.7
> 7291504
Length=569
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 54/95 (56%), Gaps = 10/95 (10%)
Query 38 LLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLL 97
++ FC +YPA S++ + + ++++ +M+W++ ++ CIETF + +
Sbjct 10 IILFCGT-LYPAYASYKAVRTKDV-------KEYVKWMMYWIVFAFFTCIETFTDI--FI 59
Query 98 QYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICP 132
+LPFYYE+K L +WL SP KG+ ++ + P
Sbjct 60 SWLPFYYEVKVALVFWLLSPATKGSSTLYRKFVHP 94
> SPCC830.08c
Length=182
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 58/117 (49%), Gaps = 10/117 (8%)
Query 36 GSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLAL 95
G LL + F PA S + E ++ K D Q + ++++ S++ IE + L
Sbjct 56 GFLLTNLLAFAMPAFFSINAI--ETTN-----KADDTQWLTYYLVTSFLNVIEYWS--QL 106
Query 96 LLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICPAYE-KAAPMCSQLYKEHCPP 151
+L Y+P Y+ +K + WLA P+F GA I+ H+I P +C + +++ P
Sbjct 107 ILYYVPVYWLLKAIFLIWLALPKFNGATIIYRHLIRPYITPHVIRICKSVSRQNAAP 163
> CE02055
Length=229
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 57/126 (45%), Gaps = 11/126 (8%)
Query 31 LGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETF 90
+ L LL +YPA S++ + + + ++++ +M+W++ + +E
Sbjct 1 MSETLSRLLIITAGTLYPAYRSYKAVRTKDT-------REYVKWMMYWIVFAIYSFLENL 53
Query 91 PPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWI---WLHVICPAYEKAAPMCSQLYKE 147
L L + PFY+++K V +WL SP KGA + W+H +EK + K
Sbjct 54 LDLVLAF-WFPFYFQLKIVFIFWLLSPWTKGASILYRKWVHPTLNRHEKDIDALLESAKS 112
Query 148 HCPPQL 153
QL
Sbjct 113 ESYNQL 118
> Hs19923919
Length=184
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 38/140 (27%), Positives = 64/140 (45%), Gaps = 11/140 (7%)
Query 18 CAGCCSIKMAFSILGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMF 77
AG ++ + + G + SLLC + FVYPA S + + SPS K+D + +
Sbjct 37 AAGAVTLLSLYLLFG-YGASLLCNLIGFVYPAYASIKAI----ESPS---KDDDTVWLTY 88
Query 78 WVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQ-FKGAGWIWLHVICPAYEK 136
WV+ + E F L LL + PFYY KC + +P+ + GA ++ V+ P + +
Sbjct 89 WVVYALFGLAEFFSDL--LLSWFPFYYVGKCAFLLFCMAPRPWNGALMLYQRVVRPLFLR 146
Query 137 AAPMCSQLYKEHCPPQLKVA 156
++ + L A
Sbjct 147 HHGAVDRIMNDLSGRALDAA 166
> At4g24960
Length=116
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 44/87 (50%), Gaps = 2/87 (2%)
Query 64 SGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAG 123
K D Q + +W++ S++ E L L++++P +Y +K V WL PQF+GA
Sbjct 14 ESTTKVDDEQWLAYWIIYSFLSLTELI--LQSLIEWIPIWYTVKLVFVAWLVLPQFQGAA 71
Query 124 WIWLHVICPAYEKAAPMCSQLYKEHCP 150
+I+ V+ ++K + S K P
Sbjct 72 FIYNRVVREQFKKHGVLRSTHSKPTKP 98
> At5g50720
Length=107
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 42/76 (55%), Gaps = 5/76 (6%)
Query 61 SSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFK 120
SPS K D Q + +W+L S++ E L LL+++P +Y K V WL PQF+
Sbjct 14 ESPS---KVDDEQWLAYWILYSFLTLSELI--LQSLLEWIPIWYTAKLVFVAWLVLPQFR 68
Query 121 GAGWIWLHVICPAYEK 136
GA +I+ V+ ++K
Sbjct 69 GAAFIYNKVVREQFKK 84
> At1g74520
Length=177
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 47/94 (50%), Gaps = 11/94 (11%)
Query 42 CVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIE-TFPPLALLLQYL 100
V VYP S Q + + + D Q + +WVL S + IE TF A L+++L
Sbjct 23 VVSLVYPLYASVQAIETQSHA-------DDKQWLTYWVLYSLLTLIELTF---AKLIEWL 72
Query 101 PFYYEMKCVLFYWLASPQFKGAGWIWLHVICPAY 134
P + MK +L WL P F GA +++ H + P +
Sbjct 73 PIWSYMKLILTCWLVIPYFSGAAYVYEHFVRPVF 106
> Hs12597657
Length=201
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 49/99 (49%), Gaps = 9/99 (9%)
Query 35 LGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLA 94
+ L+ +YPA S+ K+ S +K ++++ +M+W++ + ETF +
Sbjct 6 ISRLVVLIFGTLYPAYYSY------KAVKSKDIK-EYVKWMMYWIIFALFTTAETFTDI- 57
Query 95 LLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICPA 133
L + PFYYE+K WL SP KG+ ++ + P
Sbjct 58 -FLCWFPFYYELKIAFVAWLLSPYTKGSSLLYRKFVHPT 95
> 7301380
Length=174
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 31/136 (22%), Positives = 51/136 (37%), Gaps = 24/136 (17%)
Query 14 PWHSCAGCCSIKMAFSILGPFLGS---------------LLCFCVCFVYPASLSFQLLLD 58
PW K + F+GS LLC + +YPA +S +
Sbjct 29 PWTKAFDSLEEKTGVERVNIFVGSILFCAFYLVFGWCAQLLCNIIGVLYPAYISIHGI-- 86
Query 59 EKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQ 118
+ K D ++ +++WV IE +P +LL +PFY+ +KC W P
Sbjct 87 -----ESSTKQDDIRWLIYWVTFGIFTVIEFYP--SLLTSMIPFYWLLKCTFLIWCMLPT 139
Query 119 FKGAGWIWLHVICPAY 134
+ + H + Y
Sbjct 140 ERNGSTLIYHKLVRPY 155
> Hs7706141
Length=152
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 75 VMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICPA 133
+M+W++ ++ ET + +L + PFY+E+K WL SP KG+ ++ + P
Sbjct 1 MMYWIVFAFFTTAETLTDI--VLSWFPFYFELKIAFVIWLLSPYTKGSSVLYRKFVHPT 57
> 7303222
Length=178
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/140 (22%), Positives = 59/140 (42%), Gaps = 14/140 (10%)
Query 9 LPRFFPWHSCAGCCSIKMAFSILGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALK 68
+ R + G C+I + F + LLC + +YPA +S + + K
Sbjct 40 VDRVNIFVGAVGLCAIYLIFG----WGAQLLCNIIGVLYPAYISIHAI-------ESSTK 88
Query 69 NDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASP-QFKGAGWIWL 127
D + +++WV IE F +LL +PFY+ +KC W P + G+ I+
Sbjct 89 QDDTKWLIYWVTFGIFTVIEFFS--SLLTSVIPFYWLLKCAFLIWCMLPTEQNGSTIIYN 146
Query 128 HVICPAYEKAAPMCSQLYKE 147
++ P + K ++ +
Sbjct 147 KLVRPYFLKHHESVDRIIDD 166
> CE24624
Length=183
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 53/117 (45%), Gaps = 10/117 (8%)
Query 16 HSCAGCCSIKMAFSILGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHV 75
H G ++ + I G L+C + FVYPA +S + + E S+ K D Q +
Sbjct 41 HLVLGVVGLQALYLIFG-HSAQLVCNFMGFVYPAYMSIKAI--ESSN-----KEDDTQWL 92
Query 76 MFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICP 132
+WV+ + + +E F ++ P Y+ K + +L P F GA ++ + P
Sbjct 93 TYWVIFAILSVVEFFS--VQIVAVFPVYWLFKSIFLLYLYLPSFLGAAKLYHRFVKP 147
> Hs20473123
Length=255
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 45/101 (44%), Gaps = 13/101 (12%)
Query 34 FLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKN--DHLQHVMFWVLCSWICCIETFP 91
+ + +YPA S++ + KN ++++ +M+W++ + IET
Sbjct 5 MISRAVVLVFGMLYPAYYSYKAV---------KTKNVKEYVRWMMYWIVFALYTVIETVA 55
Query 92 PLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICP 132
+ + P YYE+K WL SP KGA I+ + P
Sbjct 56 DQTVA--WFPLYYELKIAFVIWLLSPYTKGASLIYRKFLHP 94
> SPBC30D10.09c
Length=217
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 40/81 (49%), Gaps = 6/81 (7%)
Query 52 SFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLF 111
+FQL +E S ++ + + + +W C + C L L ++PFY K V +
Sbjct 85 AFQLRNEEHKS----IEEERRRLMAYW--CVYGCVTAAESILGRFLSWVPFYSTSKIVFW 138
Query 112 YWLASPQFKGAGWIWLHVICP 132
WL +P+ +GA +I+ I P
Sbjct 139 LWLLNPRTQGAAFIYASYISP 159
> Hs20149695
Length=257
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 69 NDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLH 128
++++ +M+W++ + E + + + PFYYE+K WL SP KGA ++
Sbjct 33 REYVRWMMYWIVFALFMAAEIV--TDIFISWFPFYYEIKMAFVLWLLSPYTKGASLLYRK 90
Query 129 VICPAYEK 136
+ P+ +
Sbjct 91 FVHPSLSR 98
> ECU09g1950
Length=179
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 24/108 (22%), Positives = 51/108 (47%), Gaps = 9/108 (8%)
Query 30 ILGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVM-FWVLCSWICCIE 88
I+ LG L+ V + P L L++ + +P K D +H++ FW++ + ++
Sbjct 40 IMATSLGPLITSTVGIIVP--LQETLVILRQVNP----KKDEAKHMLVFWMVFGILTSLD 93
Query 89 TFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICPAYEK 136
+ ++ ++P +Y MK W +F+G I+ +++ EK
Sbjct 94 AYS--GAIISFIPLWYTMKFFFLLWAGPLKFRGGIIIYDNILARIPEK 139
> At1g69700
Length=184
Score = 35.4 bits (80), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 47/104 (45%), Gaps = 11/104 (10%)
Query 43 VCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIE-TFPPLALLLQYLP 101
V VYP S + + + +S P Q + +WVL + I E TF + L++ P
Sbjct 26 VTLVYPLYASVKAI-ETRSLPEDE------QWLTYWVLYALISLFELTF---SKPLEWFP 75
Query 102 FYYEMKCVLFYWLASPQFKGAGWIWLHVICPAYEKAAPMCSQLY 145
+ MK WL PQF GA I+ H I P Y ++++
Sbjct 76 IWPYMKLFGICWLVLPQFNGAEHIYKHFIRPFYRDPQRATTKIW 119
> At4g36720
Length=227
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 20/85 (23%), Positives = 37/85 (43%), Gaps = 3/85 (3%)
Query 60 KSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQF 119
K+ SG +N+ + +++W +E F ++ + P YY +K WL P
Sbjct 55 KAIESGD-ENEQQKMLIYWAAYGSFSLVEVFTDK--IISWFPLYYHVKFAFLVWLQLPTV 111
Query 120 KGAGWIWLHVICPAYEKAAPMCSQL 144
+G+ I+ + I P + QL
Sbjct 112 EGSKQIYNNQIRPFLLRHQARVDQL 136
> 7295524
Length=1713
Score = 32.3 bits (72), Expect = 0.38, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 35/67 (52%), Gaps = 2/67 (2%)
Query 31 LGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETF 90
LG L S LCFC+CFV +S+ L+ E+ S + L+ V + L +IC ++
Sbjct 1085 LGTQLASNLCFCMCFV--SSIYILFLIKERESRAKLLQFVGGVKVWTFWLSQFICDFASY 1142
Query 91 PPLALLL 97
AL++
Sbjct 1143 IVTALIV 1149
> CE17887
Length=205
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 49/107 (45%), Gaps = 9/107 (8%)
Query 26 MAFSILGPFLGSLLCFCVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWIC 85
+AF ++ + LLC + F YP S + + SP G +D +++W + +
Sbjct 90 IAFFLVFGSVARLLCNLIGFGYPTYASVKAI----RSPGG---DDDTVWLIYWTCFAVLY 142
Query 86 CIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIWLHVICP 132
++ F +L + PFYY K +L PQ +G+ + ++ P
Sbjct 143 LVDFFSEA--ILSWFPFYYIAKACFLVYLYLPQTQGSVMFYETIVDP 187
> At5g62490
Length=167
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 11/90 (12%)
Query 42 CVCFVYPASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIE-TFPPLALLLQYL 100
+ VYP S + + +S G D Q + +W L S I E TF LL+++
Sbjct 23 VISLVYPLYASVRAI---ESRSHG----DDKQWLTYWALYSLIKLFELTF---FRLLEWI 72
Query 101 PFYYEMKCVLFYWLASPQFKGAGWIWLHVI 130
P Y K L WL P GA +++ H +
Sbjct 73 PLYPYAKLALTSWLVLPGMNGAAYLYEHYV 102
> CE02157
Length=583
Score = 31.2 bits (69), Expect = 0.78, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 0/56 (0%)
Query 71 HLQHVMFWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLFYWLASPQFKGAGWIW 126
HL + FW + +CC+ T+ +L+ P Y +LF+ F + IW
Sbjct 82 HLGMICFWDIIYLLCCLSTYCIPSLIYSVTPIYGPFSYILFFLQPFASFCVSCTIW 137
> At2g17930
Length=3795
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query 49 ASLSFQLLLDEKSSPSGALKNDHLQHVMFWVLCSWICCIETFPPLALLLQ 98
A + + L D + P G + + HL V WV SWI P L L LQ
Sbjct 3059 ARVLYLLSFDTANEPVGKVFDKHLDQVPHWVWLSWI------PQLLLSLQ 3102
> Hs5174387
Length=865
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 13/24 (54%), Gaps = 0/24 (0%)
Query 21 CCSIKMAFSILGPFLGSLLCFCVC 44
CC + + F IL P +G C C C
Sbjct 112 CCVLGLLFIILMPLVGYFFCMCRC 135
> At3g55100
Length=662
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 27/56 (48%), Gaps = 3/56 (5%)
Query 77 FWVLCSWICCIETFPPLALLLQYLPFYYEMKCVLF--YWLASPQFKGAGWIWLHVI 130
FW CS++ + P ++ + F Y C+LF +++ + WIW+H I
Sbjct 497 FWSGCSFVTFVSGVIPNVMMSYMVTFGYLSYCLLFSGFYVNRDRIH-LYWIWIHYI 551
> ECU11g1080
Length=233
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 9/74 (12%)
Query 60 KSSPSGALKN---DHLQHVMFWVLCSWICCIETFPPLA-LLLQYLP--FYYEMKCVLFYW 113
KSS L N H H +FW + +C T P++ LL+Y+ F + K V +
Sbjct 74 KSSARRDLLNFSGGHYNHSLFWKM---MCPPGTSGPMSPRLLEYIERSFGSQEKMVKEFS 130
Query 114 LASPQFKGAGWIWL 127
A+ G+GW+WL
Sbjct 131 NAAVSLFGSGWVWL 144
Lambda K H
0.332 0.144 0.529
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40