bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3199_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
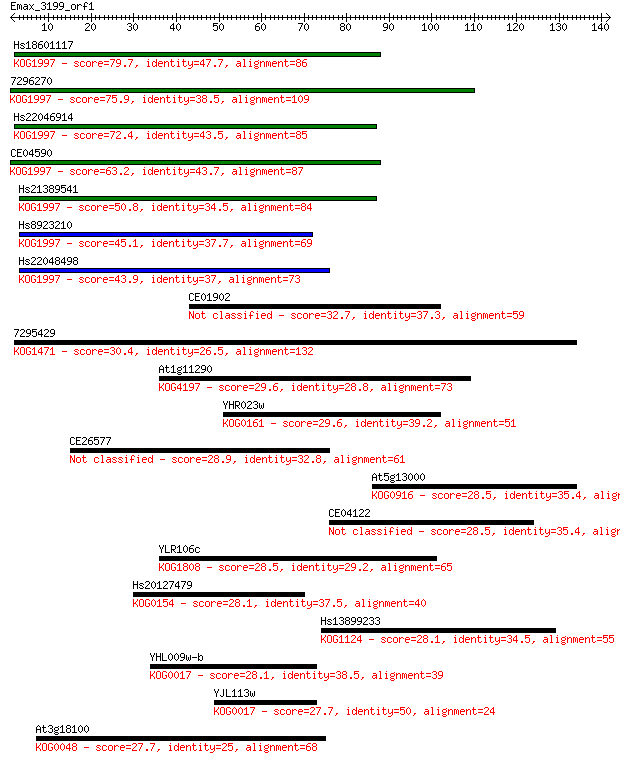
Hs18601117 79.7 2e-15
7296270 75.9 2e-14
Hs22046914 72.4 3e-13
CE04590 63.2 2e-10
Hs21389541 50.8 7e-07
Hs8923210 45.1 5e-05
Hs22048498 43.9 1e-04
CE01902 32.7 0.24
7295429 30.4 1.2
At1g11290 29.6 1.7
YHR023w 29.6 1.8
CE26577 28.9 3.1
At5g13000 28.5 4.6
CE04122 28.5 4.7
YLR106c 28.5 4.8
Hs20127479 28.1 5.5
Hs13899233 28.1 6.1
YHL009w-b 28.1 6.2
YJL113w 27.7 6.4
At3g18100 27.7 7.8
> Hs18601117
Length=2047
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/86 (47%), Positives = 53/86 (61%), Gaps = 7/86 (8%)
Query 2 TVNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGH 61
TVNQGP E+A VFL + P P + H KLR FKDFC KC +AL KN+ L+
Sbjct 1941 TVNQGPLEVAQVFLAEIPEDPKLFR-----HHNKLRLCFKDFCKKCEDALRKNKALIG-- 1993
Query 62 EQQVEYHKELERNFYKFTERLAPVMS 87
Q EYH+ELERN+ + E L P+++
Sbjct 1994 PDQKEYHRELERNYCRLREALQPLLT 2019
> 7296270
Length=1782
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 42/109 (38%), Positives = 62/109 (56%), Gaps = 6/109 (5%)
Query 1 TTVNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSG 60
TTVNQGP E+A VFL + ++ P HQ KLR F++F +C +AL KNR L+
Sbjct 1672 TTVNQGPMEMASVFLSNL----SDGTTVPTKHQNKLRLCFREFSKRCADALKKNRNLIL- 1726
Query 61 HEQQVEYHKELERNFYKFTERLAPVMSNGNVKQAIISLRESYFSGFKPI 109
Q +Y +ELERN +F ERL P ++ +V+ + +Y + P+
Sbjct 1727 -SDQKDYQRELERNNDRFIERLTPFITLTSVQNHGVVKANAYNNKSTPL 1774
> Hs22046914
Length=1573
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 53/85 (62%), Gaps = 7/85 (8%)
Query 2 TVNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGH 61
TVNQGP E+A VFL + P P + H KLR FK+F +CGEA+ KN++L++
Sbjct 1458 TVNQGPLEVAQVFLAEIPADPKLYR-----HHNKLRLCFKEFIMRCGEAVEKNKRLITA- 1511
Query 62 EQQVEYHKELERNFYKFTERLAPVM 86
Q EY +EL++N+ K E L P++
Sbjct 1512 -DQREYQQELKKNYNKLKENLRPMI 1535
> CE04590
Length=2018
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/87 (43%), Positives = 48/87 (55%), Gaps = 4/87 (4%)
Query 1 TTVNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSG 60
TTVNQGP EIA VFL + M + RP Q KLR +F+ K EA+ RQL+
Sbjct 1919 TTVNQGPLEIANVFLANA--MLDDRGRPVDRLQNKLRLSFRHLQCKAMEAIELCRQLIG- 1975
Query 61 HEQQVEYHKELERNFYKFTERLAPVMS 87
E Q EY + +E NF F L P++S
Sbjct 1976 -EDQKEYQRNVEENFESFVTHLKPMLS 2001
> Hs21389541
Length=881
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 42/84 (50%), Gaps = 7/84 (8%)
Query 3 VNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGHE 62
VN GP A FL D + A + PP +L + F+ F C AL N +L+ E
Sbjct 763 VNAGPLAYARAFLND-----SQASKYPPKKVSELEDMFRKFIQACSIALELNERLIK--E 815
Query 63 QQVEYHKELERNFYKFTERLAPVM 86
QVEYH+ L+ NF + L+ ++
Sbjct 816 DQVEYHEGLKSNFRDMVKELSDII 839
> Hs8923210
Length=500
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 26/69 (37%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 3 VNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGHE 62
VN GP A FL + NAK+ P + + L+ F+ F CG+AL N +L+ E
Sbjct 425 VNAGPMAYARAFLEE-----TNAKKYPDNQVKLLKEIFRQFADACGQALDVNERLIK--E 477
Query 63 QQVEYHKEL 71
Q+EY +EL
Sbjct 478 DQLEYQEEL 486
> Hs22048498
Length=439
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 36/73 (49%), Gaps = 7/73 (9%)
Query 3 VNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGHE 62
VN GP A FL D + A + PP +L++ F+ C AL N L+ E
Sbjct 369 VNAGPLAYAGAFLND-----SQASKYPPRKVSELKDMFRKSIQACSIALELNEWLIK--E 421
Query 63 QQVEYHKELERNF 75
QVEYH+ L+ NF
Sbjct 422 DQVEYHEGLKSNF 434
> CE01902
Length=94
Score = 32.7 bits (73), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 43 FCTKCGEALTKNRQLVSGHEQQVEYHKELERN---FYKF-TERLAPVMSNGNVKQAIISL 98
C L KNR+++ E EY K+ RN F KF T L+ S + K I+ L
Sbjct 17 LCYLAASLLQKNRKIIWIEETSTEYEKDTVRNDDLFRKFETSELSDSSSKLDDKDRIVRL 76
Query 99 RES 101
R+S
Sbjct 77 RKS 79
> 7295429
Length=324
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 35/141 (24%), Positives = 51/141 (36%), Gaps = 18/141 (12%)
Query 2 TVNQGPAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGH 61
N P E A +F + ++P K + DF + KNR ++ G
Sbjct 175 VTNWNPTEFARIFKWGEQSLPMRHKEIHLINVPSTLKWLIDFVKNRVSSKMKNRLIIYGS 234
Query 62 EQQVEYHKELERNFYKFTERLAPVMSNGNV---------KQAIISLRESYFSGFKPILSQ 112
E KEL ++ + P+ G V KQ + + R+ K IL
Sbjct 235 E------KELMKSV---DQGCLPLEMGGKVPMREMIELWKQELATKRDLILGLDKSILRS 285
Query 113 GNSVDSASSSATGKASNGGMN 133
+ SS GKAS GG N
Sbjct 286 DRGIQRRSSFNAGKASTGGPN 306
> At1g11290
Length=809
Score = 29.6 bits (65), Expect = 1.7, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 31/73 (42%), Gaps = 10/73 (13%)
Query 36 LRNAFKDFCTKCGEALTKNRQLVSGHEQQVEYHKELERNFYKFTERLAPVMSNGNVKQAI 95
+ A D KCG +L RQL G LERN + + + N N K+A+
Sbjct 273 ISTALVDMYAKCG-SLETARQLFDGM---------LERNVVSWNSMIDAYVQNENPKEAM 322
Query 96 ISLRESYFSGFKP 108
+ ++ G KP
Sbjct 323 LIFQKMLDEGVKP 335
> YHR023w
Length=1928
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 5/51 (9%)
Query 51 LTKNRQLVSGHEQQVEYHKELERNFYKFTERLAPVMSNGNVKQAIISLRES 101
+ KNR +S +E+++ Y+K LE YK E L SNG + Q + LR+S
Sbjct 1401 IIKNRDSISKYEEEIRYYK-LEN--YKLQEILNE--SNGKLSQLTLDLRQS 1446
> CE26577
Length=197
Score = 28.9 bits (63), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Query 15 LGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGHEQQVEYH--KELE 72
+GD NAK PPP+ +KL K + TK + + K R V G+ +++ +E+
Sbjct 99 IGDASQQERNAK-PPPT-AEKLLTNLKFYTTKMFQVICKCRTFVFGYFPAIDFFFPEEVA 156
Query 73 RNF 75
R F
Sbjct 157 RVF 159
> At5g13000
Length=1963
Score = 28.5 bits (62), Expect = 4.6, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 86 MSNGNVKQA--IISLRESYFSGFKPILSQGNSVDSASSSATGKASNGGMN 133
++ G V +A +I+L E F+GF L +GN V GK + G+N
Sbjct 1475 LTRGGVSKASKVINLSEDIFAGFNSTLREGN-VTHHEYIQVGKGRDVGLN 1523
> CE04122
Length=693
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 76 YKF---TERLAPVMSNGNVKQAIISLRESYFSGFKPILSQGNSVDSASSSA 123
YKF T L VMSN + ++++ SL + + S+GN +D SS+
Sbjct 408 YKFELLTRHLMRVMSNWDAQESMSSLNSDSWMSTSVVSSKGNHLDVFDSSS 458
> YLR106c
Length=4910
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 28/65 (43%), Gaps = 2/65 (3%)
Query 36 LRNAFKDFCTKCGEALTKNRQLVSGHEQQVEYHKELERNFYKFTERLAPVMSNGNVKQAI 95
LRN F + E +VS + +E K+L KF+E + GN +
Sbjct 1554 LRNRFTEIWVPSMEDFNDVNMIVS--SRLLEDLKDLANPIVKFSEWFGKKLGGGNATSGV 1611
Query 96 ISLRE 100
ISLR+
Sbjct 1612 ISLRD 1616
> Hs20127479
Length=930
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 6/46 (13%)
Query 30 PSHQQKLRNAFKDFCTKC------GEALTKNRQLVSGHEQQVEYHK 69
P ++KL + K C C EAL +++QL H+Q +E H+
Sbjct 747 PEREEKLTDWQKLACLLCRRQFPSKEALIRHQQLSGLHKQNLEIHR 792
> Hs13899233
Length=672
Score = 28.1 bits (61), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 74 NFYKFTERLAPVMSNGNVKQAIISLRESYFSGFKPILSQGNSVDSASSSATGKAS 128
NF K ER A ++ N +QAIIS + G L + N V++ + G+AS
Sbjct 459 NFEKALER-AKLVHNNEAQQAIISALDDANKGIIRELRKTNYVENLKEKSEGEAS 512
> YHL009w-b
Length=1802
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 6/39 (15%)
Query 34 QKLRNAFKDFCTKCGEALTKNRQLVSGHEQQVEYHKELE 72
QK+R + + EA++KN L HE + YHKEL+
Sbjct 1265 QKIRAIYYN------EAISKNPDLKEKHEYKQAYHKELQ 1297
> YJL113w
Length=1803
Score = 27.7 bits (60), Expect = 6.4, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 49 EALTKNRQLVSGHEQQVEYHKELE 72
EA++KN L HE + YHKEL+
Sbjct 1275 EAISKNPDLKEKHEYKQAYHKELQ 1298
> At3g18100
Length=791
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 31/70 (44%), Gaps = 2/70 (2%)
Query 7 PAEIAFVFLGDQPNMPANAKRPPPSHQQKLRNAFKDFCTKCGEALTKNRQLVSGHEQQ-- 64
P +A ++A+R P ++ L+N D C + E + +N G E+
Sbjct 636 PDSVALKKKRKAKQKKSDAERQPKRRRKGLKNCSGDVCRQENETVCENEPNNGGEERMLA 695
Query 65 VEYHKELERN 74
+E H E++ N
Sbjct 696 LECHDEIQDN 705
Lambda K H
0.314 0.130 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40