bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3161_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
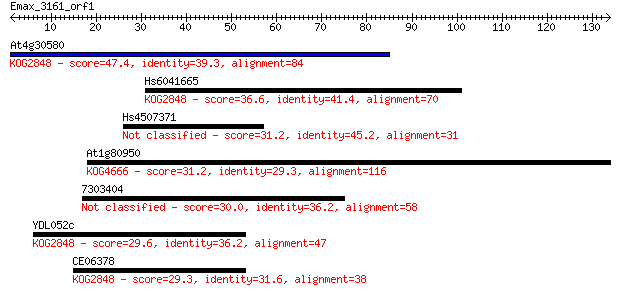
At4g30580 47.4 7e-06
Hs6041665 36.6 0.014
Hs4507371 31.2 0.55
At1g80950 31.2 0.59
7303404 30.0 1.4
YDL052c 29.6 1.6
CE06378 29.3 2.3
> At4g30580
Length=212
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 45/85 (52%), Gaps = 5/85 (5%)
Query 1 ISQLWARCAFTCAYINPNIYGKENIKGIKGPCLIISNHSSVSDIPLLGGFLPL-QHIRFV 59
I++LWA + Y NI G EN+ P + +SNH S DI L L L + +F+
Sbjct 22 IAKLWASISIYPFY-KINIEGLENLPSSDTPAVYVSNHQSFLDIYTL---LSLGKSFKFI 77
Query 60 SKAEILKWPIVGHAMREIGVVGFER 84
SK I PI+G AM +GVV +R
Sbjct 78 SKTGIFVIPIIGWAMSMMGVVPLKR 102
> Hs6041665
Length=278
Score = 36.6 bits (83), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 9/78 (11%)
Query 31 PCLIISNHSSVSDIPLLGGFLPLQHIRFVSKAEILKWPIVGHAMREIGV--VGFERNSLS 88
PC+I+SNH S+ D+ L LP + ++ ++K E+L VG M GV + +R+S +
Sbjct 91 PCVIVSNHQSILDMMGLMEVLPERCVQ-IAKRELLFLGPVGLIMYLGGVFFINRQRSSTA 149
Query 89 GTV------RLVREMFKV 100
TV R+VRE KV
Sbjct 150 MTVMADLGERMVRENLKV 167
> Hs4507371
Length=292
Score = 31.2 bits (69), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 26 KGIKGPCLIISNHSSVSDIPLLGGFLPLQHI 56
+G P + +SNH S D P L G L L+HI
Sbjct 57 RGPATPLITVSNHQSCMDDPHLWGILKLRHI 87
> At1g80950
Length=398
Score = 31.2 bits (69), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 50/121 (41%), Gaps = 26/121 (21%)
Query 18 NIYGKENIKGIKGPCLIISNHSSVSDIPLLGGFLPLQHIR-----FVSKAEILKWPIVGH 72
N G+ + + P I+SNH S DI L H+ FV+K + K P+VG
Sbjct 174 NQKGEAATEEPERPGAIVSNHVSYLDI--------LYHMSASFPSFVAKRSVGKLPLVGL 225
Query 73 AMREIGVVGFERNSLSGTVRLVREMFKVVGFRKKKEEKEKEYNKTLNPNLGFPVFLGFPE 132
+ +G V +R + S FK V + +E NK+ P + FPE
Sbjct 226 ISKCLGCVYVQREAKSPD-------FKGVSGTVNERVREAHSNKSA------PTIMLFPE 272
Query 133 G 133
G
Sbjct 273 G 273
> 7303404
Length=376
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 9/64 (14%)
Query 17 PNIYGKENI-----KGIKG-PCLIISNHSSVSDIPLLGGFLPLQHIRFVSKAEILKWPIV 70
P +Y +E + K KG P + +SNH S D P L G LPL V ++W +
Sbjct 159 PRVYNRERLIQLITKRPKGIPLVTVSNHYSCFDDPGLWGCLPLG---IVCNTYKIRWSMA 215
Query 71 GHAM 74
H +
Sbjct 216 AHDI 219
> YDL052c
Length=303
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 6 ARCAFTCAYI----NPNIYGKENIKGIKGPCLIISNHSSVSDIPLLGGFLP 52
ARC + + + + G+EN+ K P ++I+NH S DI +LG P
Sbjct 48 ARCFYHVMKLMLGLDVKVVGEENLA--KKPYIMIANHQSTLDIFMLGRIFP 96
> CE06378
Length=282
Score = 29.3 bits (64), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 21/38 (55%), Gaps = 1/38 (2%)
Query 15 INPNIYGKENIKGIKGPCLIISNHSSVSDIPLLGGFLP 52
++ +YG E + ++GP ++I NH S DI + P
Sbjct 76 VHTTVYGYEKTQ-VEGPAVVICNHQSSLDILSMASIWP 112
Lambda K H
0.323 0.143 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40