bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3134_orf3
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
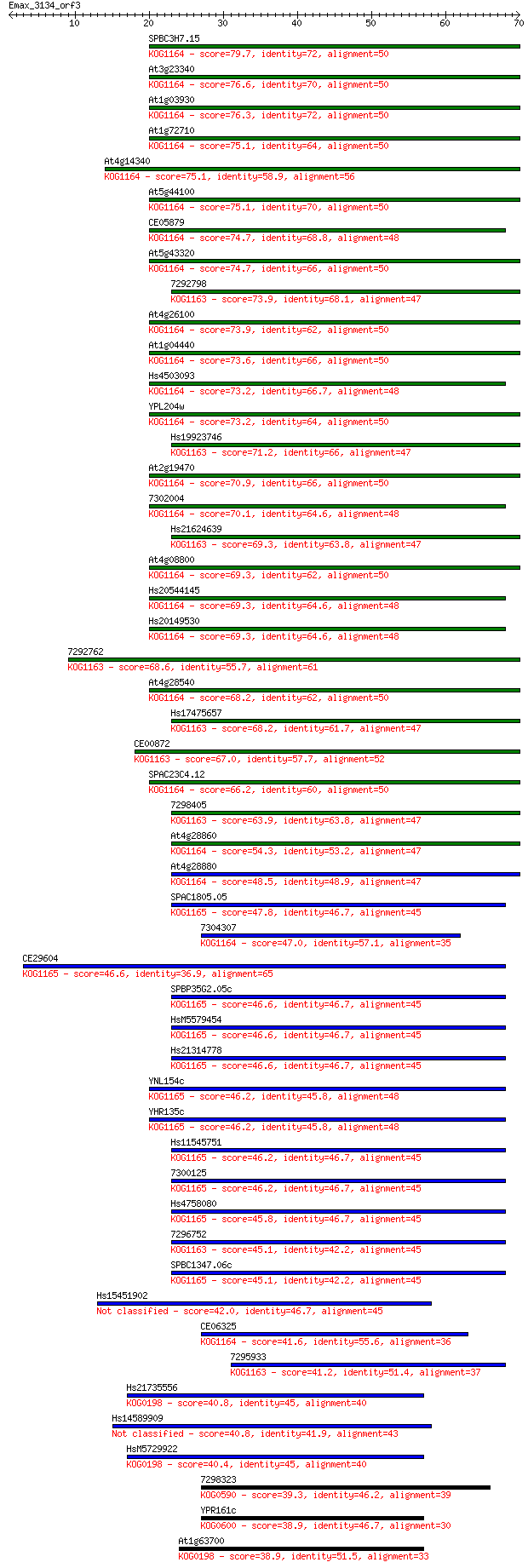
SPBC3H7.15 79.7 1e-15
At3g23340 76.6 1e-14
At1g03930 76.3 1e-14
At1g72710 75.1 3e-14
At4g14340 75.1 3e-14
At5g44100 75.1 3e-14
CE05879 74.7 4e-14
At5g43320 74.7 4e-14
7292798 73.9 6e-14
At4g26100 73.9 7e-14
At1g04440 73.6 8e-14
Hs4503093 73.2 1e-13
YPL204w 73.2 1e-13
Hs19923746 71.2 5e-13
At2g19470 70.9 6e-13
7302004 70.1 9e-13
Hs21624639 69.3 2e-12
At4g08800 69.3 2e-12
Hs20544145 69.3 2e-12
Hs20149530 69.3 2e-12
7292762 68.6 3e-12
At4g28540 68.2 4e-12
Hs17475657 68.2 4e-12
CE00872 67.0 8e-12
SPAC23C4.12 66.2 1e-11
7298405 63.9 6e-11
At4g28860 54.3 5e-08
At4g28880 48.5 3e-06
SPAC1805.05 47.8 5e-06
7304307 47.0 8e-06
CE29604 46.6 1e-05
SPBP35G2.05c 46.6 1e-05
HsM5579454 46.6 1e-05
Hs21314778 46.6 1e-05
YNL154c 46.2 1e-05
YHR135c 46.2 1e-05
Hs11545751 46.2 1e-05
7300125 46.2 1e-05
Hs4758080 45.8 2e-05
7296752 45.1 3e-05
SPBC1347.06c 45.1 3e-05
Hs15451902 42.0 3e-04
CE06325 41.6 4e-04
7295933 41.2 5e-04
Hs21735556 40.8 7e-04
Hs14589909 40.8 7e-04
HsM5729922 40.4 9e-04
7298323 39.3 0.002
YPR161c 38.9 0.002
At1g63700 38.9 0.002
> SPBC3H7.15
Length=365
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 36/50 (72%), Positives = 42/50 (84%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D RIG +R+GRKIGSGSFGDIY G NV +GEEVA+K+ES +AKHPQL Y
Sbjct 4 DLRIGNKYRIGRKIGSGSFGDIYLGTNVVSGEEVAIKLESTRAKHPQLEY 53
> At3g23340
Length=442
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/50 (70%), Positives = 42/50 (84%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D IGG ++LGRKIGSGSFG++Y G+NVQTGEEVA+K+E K KHPQL Y
Sbjct 2 DHVIGGKFKLGRKIGSGSFGELYIGINVQTGEEVALKLEPVKTKHPQLHY 51
> At1g03930
Length=471
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 36/50 (72%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D IGG ++LGRKIGSGSFG++Y G+NVQTGEEVA+K+ES K KHPQL Y
Sbjct 2 DLVIGGKFKLGRKIGSGSFGELYLGINVQTGEEVAVKLESVKTKHPQLHY 51
> At1g72710
Length=465
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 32/50 (64%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+ R+G +RLGRKIG GSFG+IY G N+QT EEVA+K+E+ K KHPQL+Y
Sbjct 2 EPRVGNKFRLGRKIGGGSFGEIYLGTNIQTNEEVAIKLENVKTKHPQLLY 51
> At4g14340
Length=457
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 33/56 (58%), Positives = 45/56 (80%), Gaps = 0/56 (0%)
Query 14 NQNPLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
++N D IGG ++LGRK+GSGSFG++Y G+N+QTGEEVA+K+E K +HPQL Y
Sbjct 2 DRNQKMDHVIGGKFKLGRKLGSGSFGELYLGINIQTGEEVAVKLEPVKTRHPQLQY 57
> At5g44100
Length=476
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 35/50 (70%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D IGG ++LG+KIGSGSFG++Y GVNVQTGEEVA+K+E+ K KHPQL Y
Sbjct 2 DLVIGGKFKLGKKIGSGSFGELYLGVNVQTGEEVAVKLENVKTKHPQLHY 51
> CE05879
Length=578
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 33/48 (68%), Positives = 40/48 (83%), Gaps = 0/48 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ R+G +RLGRKIGSGSFGDIY G N+QT EEVA+K+E K+KHPQL
Sbjct 181 ELRVGNRFRLGRKIGSGSFGDIYLGQNIQTNEEVAVKLECVKSKHPQL 228
> At5g43320
Length=480
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 33/50 (66%), Positives = 44/50 (88%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D +GG ++LGRK+GSGSFG+++ GVNVQTGEEVA+K+E A+A+HPQL Y
Sbjct 2 DRVVGGKYKLGRKLGSGSFGELFLGVNVQTGEEVAVKLEPARARHPQLHY 51
> 7292798
Length=337
Score = 73.9 bits (180), Expect = 6e-14, Method: Composition-based stats.
Identities = 32/47 (68%), Positives = 43/47 (91%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+GG +R+ RKIGSGSFGDIY G+++Q+GEEVA+K+ESA A+HPQL+Y
Sbjct 16 VGGKYRVIRKIGSGSFGDIYLGMSIQSGEEVAIKMESAHARHPQLLY 62
> At4g26100
Length=450
Score = 73.9 bits (180), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 31/50 (62%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+ R+G +RLGRKIGSGSFG+IY G N+ T EE+A+K+E+ K KHPQL+Y
Sbjct 2 EPRVGNKFRLGRKIGSGSFGEIYLGTNIHTNEELAIKLENVKTKHPQLLY 51
> At1g04440
Length=468
Score = 73.6 bits (179), Expect = 8e-14, Method: Composition-based stats.
Identities = 33/50 (66%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D +GG ++LGRK+GSGSFG+I+ GVNVQTGEEVA+K+E +A+HPQL Y
Sbjct 2 DRVVGGKFKLGRKLGSGSFGEIFLGVNVQTGEEVAVKLEPLRARHPQLHY 51
> Hs4503093
Length=416
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 32/48 (66%), Positives = 39/48 (81%), Gaps = 0/48 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ R+G +RLGRKIGSGSFGDIY G N+ +GEEVA+K+E K KHPQL
Sbjct 2 ELRVGNKYRLGRKIGSGSFGDIYLGANIASGEEVAIKLECVKTKHPQL 49
> YPL204w
Length=494
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/50 (64%), Positives = 42/50 (84%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D R+G +R+GRKIGSGSFGDIY G N+ +GEEVA+K+ES +++HPQL Y
Sbjct 2 DLRVGRKFRIGRKIGSGSFGDIYHGTNLISGEEVAIKLESIRSRHPQLDY 51
> Hs19923746
Length=337
Score = 71.2 bits (173), Expect = 5e-13, Method: Composition-based stats.
Identities = 31/47 (65%), Positives = 40/47 (85%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+GG ++L RKIGSGSFGDIY +N+ GEEVA+K+ES KA+HPQL+Y
Sbjct 13 VGGKYKLVRKIGSGSFGDIYLAINITNGEEVAVKLESQKARHPQLLY 59
> At2g19470
Length=433
Score = 70.9 bits (172), Expect = 6e-13, Method: Composition-based stats.
Identities = 33/50 (66%), Positives = 40/50 (80%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+ R+G +RLGRKIGSGSFG+IY G +VQT EEVA+K+ES K HPQL Y
Sbjct 2 EPRVGNKFRLGRKIGSGSFGEIYLGTDVQTNEEVAIKLESVKTAHPQLSY 51
> 7302004
Length=440
Score = 70.1 bits (170), Expect = 9e-13, Method: Composition-based stats.
Identities = 31/48 (64%), Positives = 38/48 (79%), Gaps = 0/48 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ R+G +RLGRKIGSGSFGDIY G + TGEEVA+K+E + KHPQL
Sbjct 2 ELRVGNKYRLGRKIGSGSFGDIYLGTTINTGEEVAIKLECIRTKHPQL 49
> Hs21624639
Length=337
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 30/47 (63%), Positives = 38/47 (80%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+GG ++L RKIGSGSFGD+Y G+ GEEVA+K+ES K KHPQL+Y
Sbjct 13 VGGKYKLVRKIGSGSFGDVYLGITTTNGEEVAVKLESQKVKHPQLLY 59
> At4g08800
Length=285
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/50 (62%), Positives = 39/50 (78%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+ RIG +RLGRKIGSG+FG+IY G +VQ+ E+VA+K ES K HPQL Y
Sbjct 2 ELRIGNKFRLGRKIGSGAFGEIYLGTDVQSNEDVAIKFESVKTVHPQLAY 51
> Hs20544145
Length=409
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/48 (64%), Positives = 38/48 (79%), Gaps = 0/48 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ R+G +RLGRKIGSGSFGDIY G ++ GEEVA+K+E K KHPQL
Sbjct 2 ELRVGNRYRLGRKIGSGSFGDIYLGTDIAAGEEVAIKLECVKTKHPQL 49
> Hs20149530
Length=415
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/48 (64%), Positives = 38/48 (79%), Gaps = 0/48 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ R+G +RLGRKIGSGSFGDIY G ++ GEEVA+K+E K KHPQL
Sbjct 2 ELRVGNRYRLGRKIGSGSFGDIYLGTDIAAGEEVAIKLECVKTKHPQL 49
> 7292762
Length=344
Score = 68.6 bits (166), Expect = 3e-12, Method: Composition-based stats.
Identities = 34/61 (55%), Positives = 46/61 (75%), Gaps = 3/61 (4%)
Query 9 ESSKSNQNPLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLI 68
E +S+QN R+ RIG +++ RKIG GSFGDIY G+ + +GE VA+KVES+K +HPQL
Sbjct 2 ERLRSSQN--REVRIGN-YKVVRKIGCGSFGDIYLGIYIHSGERVAIKVESSKVRHPQLN 58
Query 69 Y 69
Y
Sbjct 59 Y 59
> At4g28540
Length=321
Score = 68.2 bits (165), Expect = 4e-12, Method: Composition-based stats.
Identities = 31/50 (62%), Positives = 40/50 (80%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D IGG ++LGRKIG GSFG+++ V++QTGEE A+K+E AK KHPQL Y
Sbjct 2 DNVIGGKFKLGRKIGGGSFGELFLAVSLQTGEEAAVKLEPAKTKHPQLHY 51
> Hs17475657
Length=337
Score = 68.2 bits (165), Expect = 4e-12, Method: Composition-based stats.
Identities = 29/47 (61%), Positives = 38/47 (80%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
+GG ++L RKIGSGSFGD+Y G+ GE+VA+K+ES K KHPQL+Y
Sbjct 13 VGGKYKLVRKIGSGSFGDVYLGITTTNGEDVAVKLESQKVKHPQLLY 59
> CE00872
Length=341
Score = 67.0 bits (162), Expect = 8e-12, Method: Composition-based stats.
Identities = 30/52 (57%), Positives = 41/52 (78%), Gaps = 0/52 (0%)
Query 18 LRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
++D + ++L RKIGSGSFGDIY +NV GEEVA+K+ES +A+HPQL+Y
Sbjct 7 VKDFIVATKYKLIRKIGSGSFGDIYVSINVTNGEEVAIKLESNRARHPQLLY 58
> SPAC23C4.12
Length=400
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 30/50 (60%), Positives = 38/50 (76%), Gaps = 0/50 (0%)
Query 20 DARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
D +IG +R+GRKIGSGSFG IY G+N GE+VA+K+E KA+H QL Y
Sbjct 5 DIKIGNKYRIGRKIGSGSFGQIYLGLNTVNGEQVAVKLEPLKARHHQLEY 54
> 7298405
Length=367
Score = 63.9 bits (154), Expect = 6e-11, Method: Composition-based stats.
Identities = 30/47 (63%), Positives = 37/47 (78%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
IGG +RL + IGSGSFGDIY G+++ G EVA+KVE AK+PQLIY
Sbjct 23 IGGKYRLVKPIGSGSFGDIYLGLSITDGSEVAIKVEKNDAKYPQLIY 69
> At4g28860
Length=307
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/47 (53%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
IGG ++LGRKIG GSFG+I+ GV E + +E++K KHPQL+Y
Sbjct 5 IGGKYKLGRKIGGGSFGEIFLGVFSLILEFCSSSLENSKTKHPQLLY 51
> At4g28880
Length=307
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/47 (48%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQLIY 69
IGG ++LGRKIG GSFG+I+ G + + E++K KHPQL+Y
Sbjct 5 IGGKYKLGRKIGGGSFGEIFLGFVFSSVLVPLVFRENSKTKHPQLLY 51
> SPAC1805.05
Length=439
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG GSFG +++GVN+ + +A+K ES K++ PQL
Sbjct 11 VGVHYRVGKKIGEGSFGMLFQGVNLINNQPIALKFESRKSEVPQL 55
> 7304307
Length=936
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 29/35 (82%), Gaps = 0/35 (0%)
Query 27 WRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAK 61
W++ RKIG G FG+IY+G ++ T E+VA+KVESA+
Sbjct 267 WKVVRKIGGGGFGEIYEGQDLITREQVALKVESAR 301
> CE29604
Length=407
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 3 TMAHHAESSKSNQNPLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKA 62
T ++ +S S N +G +++G+KIG G+FG++ G N+ E VA+K+E K+
Sbjct 4 TRGSNSATSASTTNSQGVLMVGPNFKVGKKIGCGNFGELRLGKNLYNNEHVAIKLEPMKS 63
Query 63 KHPQL 67
K PQL
Sbjct 64 KAPQL 68
> SPBP35G2.05c
Length=435
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+GRKIG GSFG I+ G+N+ + +A+K E K++ PQL
Sbjct 8 VGVHYRVGRKIGEGSFGVIFDGMNLLNNQLIAIKFEPKKSEAPQL 52
> HsM5579454
Length=415
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG G+FG++ G N+ T E VA+K+E K++ PQL
Sbjct 42 VGPNFRVGKKIGCGNFGELRLGKNLYTNEYVAIKLEPIKSRAPQL 86
> Hs21314778
Length=415
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG G+FG++ G N+ T E VA+K+E K++ PQL
Sbjct 42 VGPNFRVGKKIGCGNFGELRLGKNLYTNEYVAIKLEPIKSRAPQL 86
> YNL154c
Length=546
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 34/49 (69%), Gaps = 1/49 (2%)
Query 20 DARIGGI-WRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
D+ I G+ +++G+KIG GSFG +++G N+ G VA+K E K + PQL
Sbjct 68 DSTIVGLHYKIGKKIGEGSFGVLFEGTNMINGLPVAIKFEPRKTEAPQL 116
> YHR135c
Length=538
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 34/49 (69%), Gaps = 1/49 (2%)
Query 20 DARIGGI-WRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
D+ I G+ +++G+KIG GSFG +++G N+ G VA+K E K + PQL
Sbjct 61 DSTIVGLHYKIGKKIGEGSFGVLFEGTNMINGVPVAIKFEPRKTEAPQL 109
> Hs11545751
Length=393
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG G+FG++ G N+ T E VA+K+E K++ PQL
Sbjct 40 VGPNFRVGKKIGCGNFGELRLGKNLYTNEYVAIKLEPIKSRAPQL 84
> 7300125
Length=422
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG G+FG++ G N+ E VA+K+E K+K PQL
Sbjct 17 VGPNFRVGKKIGCGNFGELRLGKNLYNNEHVAIKMEPMKSKAPQL 61
> Hs4758080
Length=447
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +R+G+KIG G+FG++ G N+ T E VA+K+E K++ PQL
Sbjct 39 VGPNFRVGKKIGCGNFGELRLGKNLYTNEYVAIKLEPMKSRAPQL 83
> 7296752
Length=477
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+ G +RL ++IG+GSFG++++ ++ E+VA+K+ES+ KHP L
Sbjct 64 VAGKYRLLKRIGNGSFGELFQAEGLKYHEKVAIKLESSTVKHPLL 108
> SPBC1347.06c
Length=446
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 23 IGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
+G +++GR+IG GSFG I++G N+ ++VA+K E ++ PQL
Sbjct 8 VGVHYKVGRRIGEGSFGVIFEGTNLLNNQQVAIKFEPRRSDAPQL 52
> Hs15451902
Length=894
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 29/45 (64%), Gaps = 6/45 (13%)
Query 13 SNQNPLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKV 57
S +NP D + L ++IGSG++GD+YK NV TGE A+KV
Sbjct 8 SRRNPQED------FELIQRIGSGTYGDVYKARNVNTGELAAIKV 46
> CE06325
Length=776
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/37 (54%), Positives = 27/37 (72%), Gaps = 1/37 (2%)
Query 27 WRLGRKIGSGSFGDIYKGVNVQT-GEEVAMKVESAKA 62
W++ KIG G FG+IY+ +VQ E VA+KVES+KA
Sbjct 20 WKIKAKIGGGGFGEIYEATDVQNHHERVAIKVESSKA 56
> 7295933
Length=319
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 19/37 (51%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 31 RKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHPQL 67
RK+GSGSFGDIY+ ++ +G VA+KVE A L
Sbjct 19 RKLGSGSFGDIYEAKHMGSGLHVALKVERKNAGQSHL 55
> Hs21735556
Length=620
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 17 PLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMK 56
P R R WRLG+ +G G+FG +Y +V TG E+A+K
Sbjct 347 PSRSPRAPTNWRLGKLLGQGAFGRVYLCYDVDTGRELAVK 386
> Hs14589909
Length=846
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 29/43 (67%), Gaps = 6/43 (13%)
Query 15 QNPLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMKV 57
+NP +D + L +++GSG++GD+YK NV TGE A+K+
Sbjct 14 RNPQQD------YELVQRVGSGTYGDVYKARNVHTGELAAVKI 50
> HsM5729922
Length=618
Score = 40.4 bits (93), Expect = 9e-04, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 17 PLRDARIGGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMK 56
P R R WRLG+ +G G+FG +Y +V TG E+A+K
Sbjct 345 PSRSPRAPTNWRLGKLLGQGAFGRVYLCYDVDTGRELAVK 384
> 7298323
Length=514
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 27 WRLGRKIGSGSFGDIYKGVNVQTGEEVAMKVESAKAKHP 65
W L + +G G++G++ +N QTGE VAMK+ K KHP
Sbjct 22 WTLAQTLGEGAYGEVKLLINRQTGEAVAMKMVDLK-KHP 59
> YPR161c
Length=657
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 24/30 (80%), Gaps = 0/30 (0%)
Query 27 WRLGRKIGSGSFGDIYKGVNVQTGEEVAMK 56
+R K+G G+FG++YKG++++T +VAMK
Sbjct 60 YREDEKLGQGTFGEVYKGIHLETQRQVAMK 89
> At1g63700
Length=883
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 24 GGIWRLGRKIGSGSFGDIYKGVNVQTGEEVAMK 56
G W+ GR +G GSFG +Y G N ++GE AMK
Sbjct 397 GSRWKKGRLLGMGSFGHVYLGFNSESGEMCAMK 429
Lambda K H
0.314 0.130 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200381448
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40