bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3127_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
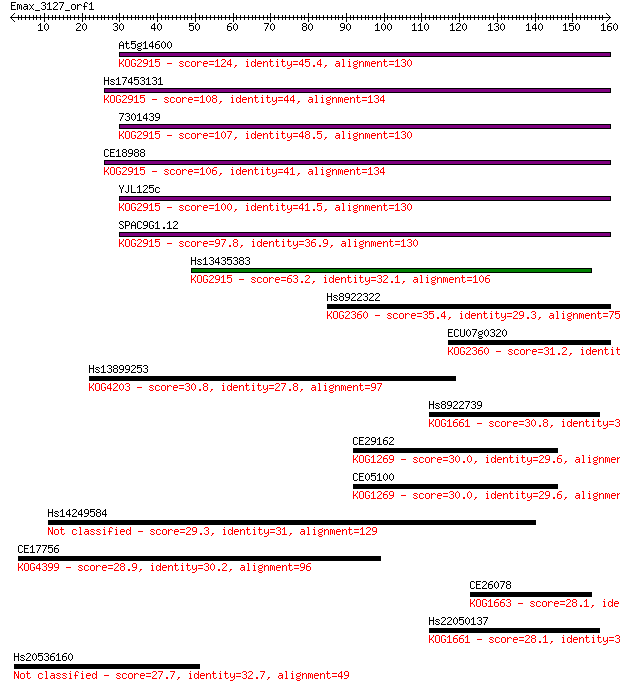
At5g14600 124 9e-29
Hs17453131 108 4e-24
7301439 107 1e-23
CE18988 106 2e-23
YJL125c 100 1e-21
SPAC9G1.12 97.8 8e-21
Hs13435383 63.2 2e-10
Hs8922322 35.4 0.050
ECU07g0320 31.2 0.86
Hs13899253 30.8 1.1
Hs8922739 30.8 1.3
CE29162 30.0 2.0
CE05100 30.0 2.0
Hs14249584 29.3 3.1
CE17756 28.9 4.5
CE26078 28.1 7.8
Hs22050137 28.1 8.0
Hs20536160 27.7 9.0
> At5g14600
Length=370
Score = 124 bits (311), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 59/130 (45%), Positives = 91/130 (70%), Gaps = 1/130 (0%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLR 89
++G+LVI+Y +++ + + G+L+ R G + H D + +G KV+ + GK++ +L
Sbjct 17 EDGDLVIVYERHDVMKPVKVSKDGVLQNRFGVYKHSDWIGKPLGTKVFSNK-GKFVYLLA 75
Query 90 PTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGR 149
PTP+L TL L HRTQI+Y ADIS ++ L+V PG ++E+GTGSGSLS SLA A+ P G
Sbjct 76 PTPELWTLVLSHRTQILYIADISFVVMYLEVVPGCVVLESGTGSGSLSTSLARAVAPTGH 135
Query 150 LFTFEFHEQR 159
+++F+FHEQR
Sbjct 136 VYSFDFHEQR 145
> Hs17453131
Length=289
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 59/135 (43%), Positives = 83/135 (61%), Gaps = 2/135 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHE-DIMKTRIGHKVYEQRQGKW 84
EE KEG+ IL +++ ++R +TR G H D++ G KV R G W
Sbjct 7 EELIKEGDTAILSLGHGAMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVTCGR-GGW 65
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMAL 144
+ VL PTP+L TL L HRTQI+Y DI+LI +L++RPG + E+GTGSGS+S+++ +
Sbjct 66 VYVLHPTPELWTLNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRTI 125
Query 145 RPGGRLFTFEFHEQR 159
P G L T EFH+QR
Sbjct 126 APTGHLHTVEFHQQR 140
> 7301439
Length=317
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 63/140 (45%), Positives = 87/140 (62%), Gaps = 12/140 (8%)
Query 30 KEGELVILYGSREMI-----ISCVLRRAG-----ILRTRLGNFSHEDIMKTRIGHKVYEQ 79
++G++VILY S + + ++ + G I +T G+ E+I+ G KV E
Sbjct 11 EKGDVVILYLSVSSMHAIEAVPEIVNKKGETIPHIFQTNYGSLKVENIIGVEYGSKV-EL 69
Query 80 RQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYS 139
+G W VL+PTP+L T L HRTQIIY DIS+I+ L+VRPG +VE+GTGSGSLS+
Sbjct 70 SKG-WAHVLQPTPELWTQTLPHRTQIIYTPDISMIIHQLEVRPGAVVVESGTGSGSLSHY 128
Query 140 LAMALRPGGRLFTFEFHEQR 159
AL+P G L TF+FHE R
Sbjct 129 FLRALKPTGHLHTFDFHEAR 148
> CE18988
Length=312
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 83/134 (61%), Gaps = 2/134 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWL 85
E+ +EG+ VI+Y + ++ +++R L + G HE I+ R G ++ ++
Sbjct 12 EDRIQEGDTVIVYVTYGQMVPTIVKRGQTLMMKYGALRHEFIIGKRWGQRL--SATAGYV 69
Query 86 VVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
+LRPT DL T AL RTQI+Y AD + ILSLLD +PG I E+GTGSGSLS+++A+ +
Sbjct 70 YILRPTSDLWTRALPRRTQILYTADCAQILSLLDAKPGSVICESGTGSGSLSHAIALTVA 129
Query 146 PGGRLFTFEFHEQR 159
P G L+T + E R
Sbjct 130 PTGHLYTHDIDETR 143
> YJL125c
Length=383
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 54/134 (40%), Positives = 83/134 (61%), Gaps = 7/134 (5%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGK----WL 85
KEG+L +++ SR+ I + + TR G+F H+DI+ G ++ + +G ++
Sbjct 14 KEGDLTLIWVSRDNIKPVRMHSEEVFNTRYGSFPHKDIIGKPYGSQIAIRTKGSNKFAFV 73
Query 86 VVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
VL+PTP+L TL+L HRTQI+Y D S I+ L+ P ++EAGTGSGS S++ A ++
Sbjct 74 HVLQPTPELWTLSLPHRTQIVYTPDSSYIMQRLNCSPHSRVIEAGTGSGSFSHAFARSV- 132
Query 146 PGGRLFTFEFHEQR 159
G LF+FEFH R
Sbjct 133 --GHLFSFEFHHIR 144
> SPAC9G1.12
Length=364
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 48/130 (36%), Positives = 78/130 (60%), Gaps = 0/130 (0%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLR 89
+ G+L + + R +I + + G F H +++ R G ++ + ++ +L+
Sbjct 11 ENGDLAVAWIGRNKLIPLHIEAEKTFHNQYGAFPHSEMIGKRYGEQIASTAKQGFIYLLQ 70
Query 90 PTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGR 149
PTP+L TLAL HRTQI+Y DI+LI L + G ++EAGTGS S+S++++ + P GR
Sbjct 71 PTPELWTLALPHRTQIVYTPDIALIHQKLRITYGTRVIEAGTGSASMSHAISRTVGPLGR 130
Query 150 LFTFEFHEQR 159
LFTFE+H R
Sbjct 131 LFTFEYHATR 140
> Hs13435383
Length=477
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 60/106 (56%), Gaps = 1/106 (0%)
Query 49 LRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYH 108
L G+L + G I+ G ++ GK ++ RP + + + ++ T I +
Sbjct 174 LNNFGLLNSNWGAVPFGKIVGKFPG-QILRSSFGKQYMLRRPALEDYVVLMKRGTAITFP 232
Query 109 ADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFE 154
DI++ILS++D+ PG T++EAG+GSG +S L+ A+ GR+ +FE
Sbjct 233 KDINMILSMMDINPGDTVLEAGSGSGGMSLFLSKAVGSQGRVISFE 278
> Hs8922322
Length=429
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILS-LLDVRPGKTIVEAGTGSGSLSYSLAMA 143
L+V DLH L +I S + + LLD PG +++A G+ + LA
Sbjct 188 LLVFPAQTDLHEHPLYRAGHLILQDRASCLPAMLLDPPPGSHVIDACAAPGNKTSHLAAL 247
Query 144 LRPGGRLFTFEFHEQR 159
L+ G++F F+ +R
Sbjct 248 LKNQGKIFAFDLDAKR 263
> ECU07g0320
Length=364
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 117 LLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQR 159
+LD G +++ + G+ + LAM +R G+++ FE + R
Sbjct 171 ILDPEEGSRVIDTCSAPGNKTSQLAMIMRNTGKIYAFERSKNR 213
> Hs13899253
Length=277
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 4/98 (4%)
Query 22 LQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQ-R 80
L Q+E ++ ++VIL R + ++A L+ + NF H D + H+ +
Sbjct 47 LGQNEVEQRQRKVVILSQDRFYKVLTAEQKAKALKGQY-NFDHPDAFDNDLMHRTLKNIV 105
Query 81 QGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLL 118
+GK + V PT D T + T ++Y AD+ L +L
Sbjct 106 EGKTVEV--PTYDFVTHSRLPETTVVYPADVVLFEGIL 141
> Hs8922739
Length=361
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ LD++PG + + G+G+G LS + + L P G E H
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILGPFGVNHGVELH 114
> CE29162
Length=484
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 92 PDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
PD +++ L H + + +D + IL+ L + K +V+ G G G + LA R
Sbjct 44 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 97
> CE05100
Length=495
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 92 PDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
PD +++ L H + + +D + IL+ L + K +V+ G G G + LA R
Sbjct 55 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 108
> Hs14249584
Length=352
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 54/129 (41%), Gaps = 19/129 (14%)
Query 11 AEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKT 70
AEE+A +SA E PK V+ + S+E S + R + T+ S K+
Sbjct 146 AEEAACPVSA---PEEASPKP---VLCHQSKERKPSAEMNR---ITTKEATSSCPP--KS 194
Query 71 RIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAG 130
+G E RQ W + L R R Q+ H + SLLDVR I G
Sbjct 195 PLG----ETRQKLWRSLKM----LPERGQRVRQQLKSHLATVNLSSLLDVRRSTVISGPG 246
Query 131 TGSGSLSYS 139
TG GS +S
Sbjct 247 TGKGSQDHS 255
> CE17756
Length=467
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 3 PPFGVFVEAEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNF 62
PPFGVF+E L + + + F G+ L+ S MI+ + R +L GNF
Sbjct 310 PPFGVFMEP-----LLKSIEKMKKRFVSTGKEETLFYS--MIVLPIYIRKYVLH---GNF 359
Query 63 SHEDIMKTRIGHKVYEQRQGKWLVVLRPTP----DLHTLA 98
D T GHK+Y+ + + + P DL +A
Sbjct 360 WMSDYRVTYEGHKLYQHSEKTIVRLFTDLPVECIDLKNVA 399
> CE26078
Length=219
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 123 GKTIVEAGTGSGSLSYSLAMALRPGGRLFTFE 154
G+ +++ GT +G+ + + A+A+ G +FTF+
Sbjct 57 GRRVIDVGTYTGASALAWALAVPDDGEVFTFD 88
> Hs22050137
Length=357
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ L ++PG + + G+G+G LS + + L P G E H
Sbjct 70 SEVMEALKLQPGLSFLNLGSGTGYLSTMVGLILGPFGINHGIELH 114
> Hs20536160
Length=215
Score = 27.7 bits (60), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 APPFGVFVEAEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLR 50
A F + + + A LQQ E+ PK +L +L+ S M +S +LR
Sbjct 167 ATNFTMAANSSNGQTKTGAPLQQGEDPPKRRDLALLHLSVFMAVSALLR 215
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40