bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3124_orf1
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
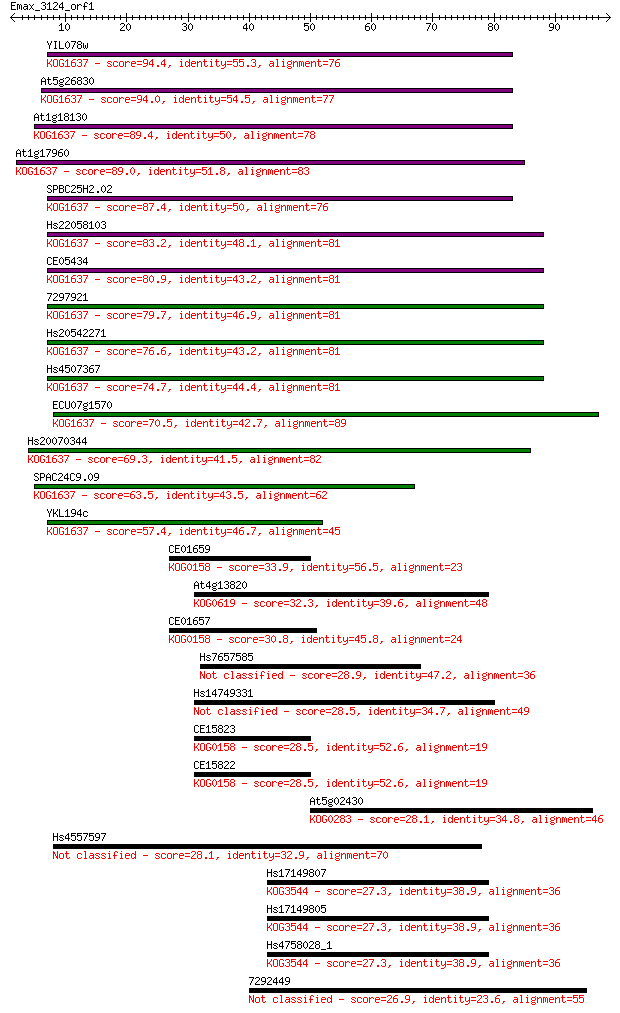
YIL078w 94.4 5e-20
At5g26830 94.0 6e-20
At1g18130 89.4 1e-18
At1g17960 89.0 2e-18
SPBC25H2.02 87.4 6e-18
Hs22058103 83.2 1e-16
CE05434 80.9 5e-16
7297921 79.7 1e-15
Hs20542271 76.6 9e-15
Hs4507367 74.7 4e-14
ECU07g1570 70.5 7e-13
Hs20070344 69.3 2e-12
SPAC24C9.09 63.5 8e-11
YKL194c 57.4 6e-09
CE01659 33.9 0.085
At4g13820 32.3 0.24
CE01657 30.8 0.58
Hs7657585 28.9 2.7
Hs14749331 28.5 3.1
CE15823 28.5 3.3
CE15822 28.5 3.3
At5g02430 28.1 3.7
Hs4557597 28.1 3.9
Hs17149807 27.3 6.2
Hs17149805 27.3 6.2
Hs4758028_1 27.3 6.2
7292449 26.9 8.9
> YIL078w
Length=734
Score = 94.4 bits (233), Expect = 5e-20, Method: Composition-based stats.
Identities = 42/76 (55%), Positives = 54/76 (71%), Gaps = 0/76 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPV+IHRAILGS+ERM A+L EH GK PFWLSPRQ +V+ + K GYAE V + L
Sbjct 594 RPVMIHRAILGSVERMTAILTEHFAGKWPFWLSPRQVLVVPVGVKYQGYAEDVRNKLHDA 653
Query 67 GYDVGVDVSNNTITRK 82
G+ VD++ NT+ +K
Sbjct 654 GFYADVDLTGNTLQKK 669
> At5g26830
Length=676
Score = 94.0 bits (232), Expect = 6e-20, Method: Composition-based stats.
Identities = 42/77 (54%), Positives = 56/77 (72%), Gaps = 0/77 (0%)
Query 6 ARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQR 65
+RPV+IHRA+LGS+ERM A+LLEH GK PFW+SPRQ IV ISEK+ YAE V ++
Sbjct 545 SRPVMIHRAVLGSVERMFAILLEHYKGKWPFWISPRQAIVCPISEKSQQYAEKVQKQIKD 604
Query 66 EGYDVGVDVSNNTITRK 82
G+ V D+++ I +K
Sbjct 605 AGFYVDADLTDRKIDKK 621
> At1g18130
Length=165
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 5 MARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQ 64
+ R V+IHRA+L S+ER+ A+LLE GK PFWLSPRQ IV S+SE YA+ V D +
Sbjct 37 IERSVLIHRAVLSSVERLFAILLEQYKGKFPFWLSPRQAIVCSVSEDYRSYADKVRDQIH 96
Query 65 REGYDVGVDVSNNTITRK 82
GY V VD ++ ++ K
Sbjct 97 EAGYYVDVDTTDRNVSDK 114
> At1g17960
Length=458
Score = 89.0 bits (219), Expect = 2e-18, Method: Composition-based stats.
Identities = 43/83 (51%), Positives = 55/83 (66%), Gaps = 0/83 (0%)
Query 2 KPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVAD 61
K M PV+IH A+LGSIERM A+LLEH G PFWLSPRQ IV S+SE + YA+ V
Sbjct 319 KNEMEAPVMIHSAVLGSIERMFAILLEHYKGIWPFWLSPRQAIVCSLSEDCSSYAKQVQK 378
Query 62 VLQREGYDVGVDVSNNTITRKSA 84
+ GY V +D S+ ++ +K A
Sbjct 379 QINEVGYYVDIDESDRSLRKKVA 401
> SPBC25H2.02
Length=703
Score = 87.4 bits (215), Expect = 6e-18, Method: Composition-based stats.
Identities = 38/76 (50%), Positives = 52/76 (68%), Gaps = 0/76 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPV++HRAILGS+ERM A+L EH GK PFW+SPRQ ++ +S A YA+ V ++L
Sbjct 564 RPVMVHRAILGSLERMIAILTEHYAGKWPFWMSPRQVCIIPVSAAAYNYADKVHNILSEA 623
Query 67 GYDVGVDVSNNTITRK 82
V D S+NT+ +K
Sbjct 624 DIFVDTDKSDNTLPKK 639
> Hs22058103
Length=707
Score = 83.2 bits (204), Expect = 1e-16, Method: Composition-based stats.
Identities = 39/81 (48%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GGK PFWLSPRQ +V+ + YA V+ E
Sbjct 569 RPVIIHRAILGSVERMIAILSENYGGKWPFWLSPRQVMVIPVGPTCEKYALQVSSEFFEE 628
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+ VD+ ++ K R++
Sbjct 629 GFMADVDLDHSCTLNKKIRNA 649
> CE05434
Length=725
Score = 80.9 bits (198), Expect = 5e-16, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPV+IHRA+LGS+ERM A+L E GGK PFWLSPRQC ++++ E YA V +
Sbjct 584 RPVMIHRAVLGSVERMTAILTESYGGKWPFWLSPRQCKIITVHESVRDYANDVKKQIFEA 643
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+++ + + K R +
Sbjct 644 GFEIEYEENCGDTMNKQVRKA 664
> 7297921
Length=690
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/81 (46%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GK PFWLSPRQ +V+ + YA++V D L
Sbjct 553 RPVIIHRAILGSVERMIAILTENFAGKWPFWLSPRQVMVVPVGPAYDQYAQSVRDQLHDA 612
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+ D K R++
Sbjct 613 GFMSEADCDAGDTMNKKIRNA 633
> Hs20542271
Length=723
Score = 76.6 bits (187), Expect = 9e-15, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 48/81 (59%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVI+HRAILGS+ERM A+L E+ GGK PFWLSPRQ +V+ + YA+ V
Sbjct 585 RPVIVHRAILGSVERMIAILTENYGGKWPFWLSPRQVMVVPVGPTCDEYAQKVRQQFHDA 644
Query 67 GYDVGVDVSNNTITRKSARHS 87
+ +D+ K R++
Sbjct 645 KFMADIDLDPGCTLNKKIRNA 665
> Hs4507367
Length=712
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 36/82 (43%), Positives = 49/82 (59%), Gaps = 1/82 (1%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKL-PFWLSPRQCIVLSISEKAAGYAETVADVLQR 65
RPVI+HRAILGS+ERM A+L E+ GGKL PFWLSPRQ +V+ + YA+ V
Sbjct 573 RPVIVHRAILGSVERMIAILTENYGGKLAPFWLSPRQVMVVPVGPTCDEYAQNVRQQFHD 632
Query 66 EGYDVGVDVSNNTITRKSARHS 87
+ +D+ K R++
Sbjct 633 AKFMADIDLDPGCTLNKKIRNA 654
> ECU07g1570
Length=640
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 38/89 (42%), Positives = 51/89 (57%), Gaps = 3/89 (3%)
Query 8 PVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQREG 67
PVIIHRAILGSIERM A++LE G +LPFW+SPRQ ++++ Y V VL R
Sbjct 502 PVIIHRAILGSIERMIAIILESFGKRLPFWISPRQIAIVNMGN--PDYVGKVRSVLARFR 559
Query 68 YDVGVDVSNNTITRKSARHSCSNGICCLL 96
DV +D N R + + C++
Sbjct 560 LDV-IDDGNTLSKRIRTAETSGYALVCVV 587
> Hs20070344
Length=718
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 4 GMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVL 63
+ RPV+IHRA+LGS+ER+ VL E GGK P WLSP Q +V+ + + YA+ L
Sbjct 577 ALERPVLIHRAVLGSVERLLGVLAESCGGKWPLWLSPFQVVVIPVGSEQEEYAKEAQQSL 636
Query 64 QREGYDVGVDV-SNNTITRKSAR 85
+ G +D S T++R+ R
Sbjct 637 RAAGLVSDLDADSGLTLSRRIRR 659
> SPAC24C9.09
Length=473
Score = 63.5 bits (153), Expect = 8e-11, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 5 MARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSI--SEKAAGYAETVADV 62
+ +PV+IHRAI GS+ER +L+EH G PFWLSPR ++L + +++ YA+ V
Sbjct 323 IKQPVMIHRAIFGSLERFMGILIEHLNGHWPFWLSPRHAVILPVNQTDRILTYAKQVQKE 382
Query 63 LQRE 66
L +
Sbjct 383 LSNQ 386
> YKL194c
Length=462
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEK 51
RP++IHRA GSIER A+L++ G+ PFWL+P Q +++ ++ K
Sbjct 314 RPIMIHRATFGSIERFMALLIDSNEGRWPFWLNPYQAVIIPVNTK 358
> CE01659
Length=515
Score = 33.9 bits (76), Expect = 0.085, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 27 LEHTGGKLPFWLSPRQCIVLSIS 49
LEH G LPF L PRQCI + ++
Sbjct 444 LEHKGAYLPFGLGPRQCIGMRLA 466
> At4g13820
Length=645
Score = 32.3 bits (72), Expect = 0.24, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 31 GGKLPFWL-SPRQCIVLSISEKAAGYAETVADVLQREGYDVGVDVSNNT 78
GG++P WL S + ++IS+ + E ADV+QR G + +D+S+NT
Sbjct 232 GGQVPQWLWSLPELQYVNISQNSFSGFEGPADVIQRCGELLMLDISSNT 280
> CE01657
Length=518
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 27 LEHTGGKLPFWLSPRQCIVLSISE 50
LEH G +PF PRQCI + +++
Sbjct 447 LEHNGSYIPFGSGPRQCIGMRLAQ 470
> Hs7657585
Length=301
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 7/43 (16%)
Query 32 GKLPFWLS--PRQCI-----VLSISEKAAGYAETVADVLQREG 67
G + WL+ P CI VLS+S K AG+ T +V++ EG
Sbjct 219 GGICLWLAVYPVDCIKSRIQVLSMSGKQAGFIRTFINVVKNEG 261
> Hs14749331
Length=229
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 3/52 (5%)
Query 31 GGKLPFWLSPRQCIVLSISEKAAGYAETVA---DVLQREGYDVGVDVSNNTI 79
G +LP WL P +C V ++ A A++V D+ + G V V+NN +
Sbjct 63 GPQLPSWLQPERCAVFQCAQCHAVLADSVHLAWDLSRSLGAVVFSRVTNNVV 114
> CE15823
Length=518
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 31 GGKLPFWLSPRQCIVLSIS 49
GG +PF L PRQCI + ++
Sbjct 450 GGYIPFGLGPRQCIGMRLA 468
> CE15822
Length=517
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 31 GGKLPFWLSPRQCIVLSIS 49
GG +PF L PRQCI + ++
Sbjct 449 GGYIPFGLGPRQCIGMRLA 467
> At5g02430
Length=905
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 50 EKAAGYAETVADVLQREGYDVGVDVSN-NTITRKSARHSCSNGICCL 95
EK+ GY+ V D+++RE + +D N+ KS R S G L
Sbjct 296 EKSVGYSSVVKDLMRRENANSTMDFRKFNSYVSKSLRVSKKRGAALL 342
> Hs4557597
Length=2705
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 32/74 (43%), Gaps = 5/74 (6%)
Query 8 PVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPR----QCIVLSISEKAAGYAETVADVL 63
P IH G + C V + G L L PR Q V+++ KAAG + V
Sbjct 1605 PFRIHALPTGDASK-CLVTVSIGGHGLGACLGPRIQIGQETVITVDAKAAGEGKVTCTVS 1663
Query 64 QREGYDVGVDVSNN 77
+G ++ VDV N
Sbjct 1664 TPDGAELDVDVVEN 1677
> Hs17149807
Length=2976
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query 43 CIVLSISEKAAGYAETVADVLQREGYDV--GVDVSNNT 78
C+ S+S + AG ET+ D+ ++ D+ +D SNNT
Sbjct 216 CVHSSVSPERAGDTETLKDITAQDSADIIFLIDGSNNT 253
> Hs17149805
Length=3009
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query 43 CIVLSISEKAAGYAETVADVLQREGYDV--GVDVSNNT 78
C+ S+S + AG ET+ D+ ++ D+ +D SNNT
Sbjct 216 CVHSSVSPERAGDTETLKDITAQDSADIIFLIDGSNNT 253
> Hs4758028_1
Length=2079
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query 43 CIVLSISEKAAGYAETVADVLQREGYDV--GVDVSNNT 78
C+ S+S + AG ET+ D+ ++ D+ +D SNNT
Sbjct 216 CVHSSVSPERAGDTETLKDITAQDSADIIFLIDGSNNT 253
> 7292449
Length=516
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 13/59 (22%), Positives = 26/59 (44%), Gaps = 4/59 (6%)
Query 40 PRQCIVLSISE----KAAGYAETVADVLQREGYDVGVDVSNNTITRKSARHSCSNGICC 94
P C S E K +G+ + +++R G D+ ++ + + +C+NG C
Sbjct 70 PNNCPWASCGEWCLTKTSGFCPQIHSIVRRNGTDIQLNNCTRIDLSRLNKFNCNNGTAC 128
Lambda K H
0.323 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194657780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40