bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3113_orf2
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
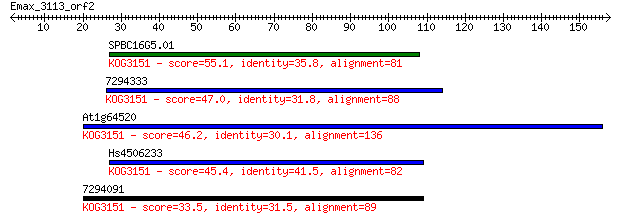
SPBC16G5.01 55.1 6e-08
7294333 47.0 2e-05
At1g64520 46.2 2e-05
Hs4506233 45.4 4e-05
7294091 33.5 0.19
> SPBC16G5.01
Length=159
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 49/83 (59%), Gaps = 5/83 (6%)
Query 27 AFQRHMNMLLPYYCDLAHVLPPSPMQQELQCLQLLHYLAGDCIAELHTAYDRLPEE--IQ 84
+F R+ + ++P+Y D L PS + L LL+ L+ + IAE HTA + +P++ +
Sbjct 69 SFARYASQVIPFYHD---SLVPSSRMGLVTGLNLLYLLSENRIAEFHTALESVPDKSLFE 125
Query 85 RSPYIVTVMRLEQQLMSGDYEAV 107
R PY+ V+ LEQ +M G ++ V
Sbjct 126 RDPYVEWVISLEQNVMEGAFDKV 148
> 7294333
Length=262
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 44/89 (49%), Gaps = 1/89 (1%)
Query 26 AAFQRHMNMLLPYYCDLAHVLPPSPMQQELQCLQLLHYLAGDCIAELHTAYDRLPEE-IQ 84
AAF+R+M L YY D L S + L LL+ LA + +A+ H +RLP +
Sbjct 74 AAFERYMAQLNTYYYDYDKYLESSKHMYKFMGLNLLYMLATNRLADFHIELERLPTALLL 133
Query 85 RSPYIVTVMRLEQQLMSGDYEAVMKQQQQ 113
+I V+ LE M G Y +++ ++
Sbjct 134 HDSFIQPVLALENYYMEGRYNKILQAKKS 162
> At1g64520
Length=267
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 57/137 (41%), Gaps = 21/137 (15%)
Query 20 MMTETAAAFQRHMNMLLPYYCDLAHVLPPSPMQQELQCLQLLHYLAGDCIAELHTAYDRL 79
+ TE AF+R L PYY D + +P SP + + L LL L + IAE HT + L
Sbjct 73 VKTEDQDAFERDFFQLKPYYVDARNRIPQSPQENLILGLNLLRLLVQNRIAEFHTELELL 132
Query 80 PEEIQRSPYIVTVMRLEQQLMSGDYEAVMKQQQQQLQQQQQQQQQQQQQQQQVCLPRSAA 139
P I + LEQ M G Y V+ +Q P +
Sbjct 133 SSATLEDPCIKHAVELEQSFMEGAYNRVLSARQTA--------------------PDATY 172
Query 140 V-FLSQLTKTVQQQIAA 155
V F+ L KT++ +IA
Sbjct 173 VYFMDLLAKTIRDEIAG 189
> Hs4506233
Length=257
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 46/83 (55%), Gaps = 1/83 (1%)
Query 27 AFQRHMNMLLPYYCDLAHVLPPSPMQQELQCLQLLHYLAGDCIAELHTAYDRLP-EEIQR 85
+F+R+M L YY D LP S +L L LL L+ + +AE HT +RLP ++IQ
Sbjct 70 SFERYMAQLKCYYFDYKEQLPESAYMHQLLGLNLLFLLSQNRVAEFHTELERLPAKDIQT 129
Query 86 SPYIVTVMRLEQQLMSGDYEAVM 108
+ YI + LEQ LM G Y V
Sbjct 130 NVYIKHPVSLEQYLMEGSYNKVF 152
> 7294091
Length=264
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 52/90 (57%), Gaps = 1/90 (1%)
Query 20 MMTETAAAFQRHMNMLLPYYCDLAHVLPPSPMQQELQCLQLLHYLAGDCIAELHTAYDRL 79
++++ AF+R+M L YY D A ++ S + +L L LL+ L+G+ +++ HT + L
Sbjct 71 VLSKDLLAFERYMAQLKCYYYDYAKIIGESESKYKLLGLNLLYLLSGNRVSDFHTELELL 130
Query 80 P-EEIQRSPYIVTVMRLEQQLMSGDYEAVM 108
+ IQ + +I ++ LEQ +M G Y +
Sbjct 131 SVDVIQHNQFIRPILALEQYIMEGRYNKIF 160
Lambda K H
0.320 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40