bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
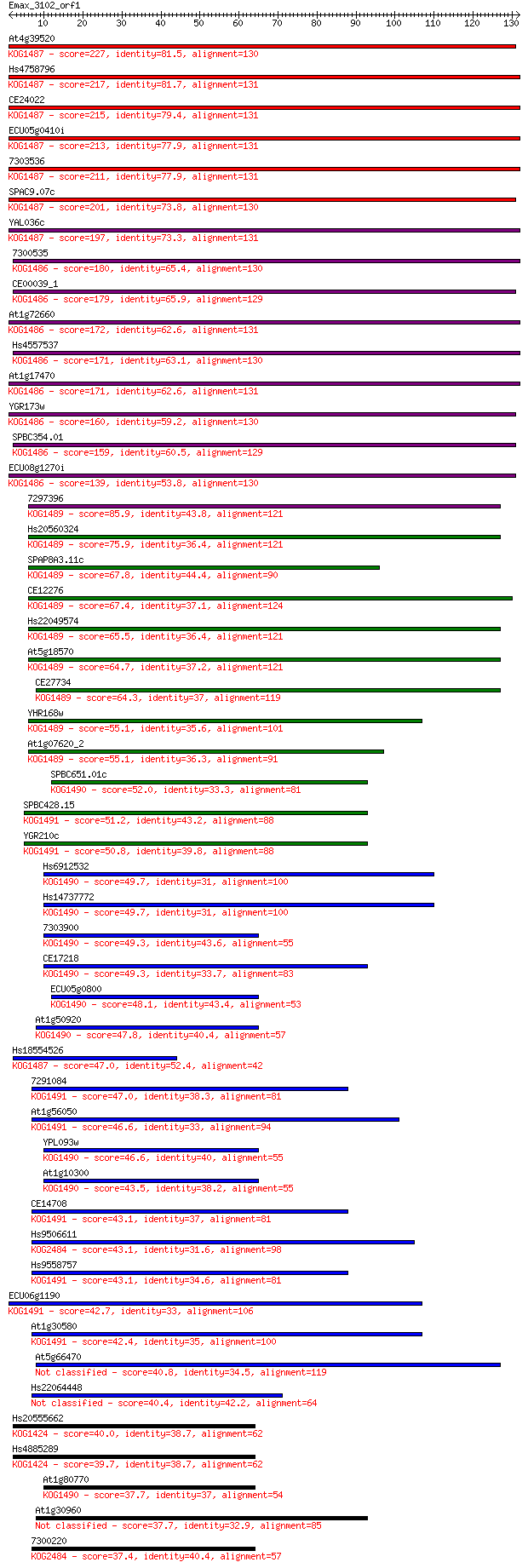
Query= Emax_3102_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
At4g39520 227 5e-60
Hs4758796 217 5e-57
CE24022 215 2e-56
ECU05g0410i 213 1e-55
7303536 211 2e-55
SPAC9.07c 201 2e-52
YAL036c 197 3e-51
7300535 180 7e-46
CE00039_1 179 9e-46
At1g72660 172 2e-43
Hs4557537 171 3e-43
At1g17470 171 4e-43
YGR173w 160 7e-40
SPBC354.01 159 1e-39
ECU08g1270i 139 1e-33
7297396 85.9 2e-17
Hs20560324 75.9 2e-14
SPAP8A3.11c 67.8 5e-12
CE12276 67.4 8e-12
Hs22049574 65.5 3e-11
At5g18570 64.7 5e-11
CE27734 64.3 6e-11
YHR168w 55.1 3e-08
At1g07620_2 55.1 3e-08
SPBC651.01c 52.0 3e-07
SPBC428.15 51.2 5e-07
YGR210c 50.8 7e-07
Hs6912532 49.7 2e-06
Hs14737772 49.7 2e-06
7303900 49.3 2e-06
CE17218 49.3 2e-06
ECU05g0800 48.1 4e-06
At1g50920 47.8 5e-06
Hs18554526 47.0 8e-06
7291084 47.0 9e-06
At1g56050 46.6 1e-05
YPL093w 46.6 1e-05
At1g10300 43.5 1e-04
CE14708 43.1 1e-04
Hs9506611 43.1 1e-04
Hs9558757 43.1 1e-04
ECU06g1190 42.7 2e-04
At1g30580 42.4 2e-04
At5g66470 40.8 7e-04
Hs22064448 40.4 8e-04
Hs20555662 40.0 0.001
Hs4885289 39.7 0.002
At1g80770 37.7 0.005
At1g30960 37.7 0.006
7300220 37.4 0.008
> At4g39520
Length=369
Score = 227 bits (578), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 106/130 (81%), Positives = 120/130 (92%), Gaps = 0/130 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+K+GD+RVGLVGFPSVGKSTLLNKLTGTFSEVA+YEFTTLTC+PGV Y+GAK+QLLDLP
Sbjct 61 TKSGDSRVGLVGFPSVGKSTLLNKLTGTFSEVASYEFTTLTCIPGVITYRGAKIQLLDLP 120
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDGKGRGRQVI ARTC+ ILIVLD + P+THK +IE+ELEGFGIRLNK+PPN+
Sbjct 121 GIIEGAKDGKGRGRQVISTARTCNCILIVLDAIKPITHKRLIEKELEGFGIRLNKEPPNL 180
Query 121 TIHKKDKGGI 130
T KKDKGGI
Sbjct 181 TFRKKDKGGI 190
> Hs4758796
Length=367
Score = 217 bits (552), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 107/131 (81%), Positives = 114/131 (87%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+KTGDAR+G VGFPSVGKSTLL+ L G +SEVAAYEFTTLT VPGV YKGAK+QLLDLP
Sbjct 60 AKTGDARIGFVGFPSVGKSTLLSNLAGVYSEVAAYEFTTLTTVPGVIRYKGAKIQLLDLP 119
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDGKGRGRQVI VARTC+LILIVLDVL PL HK IIE ELEGFGIRLN +PPNI
Sbjct 120 GIIEGAKDGKGRGRQVIAVARTCNLILIVLDVLKPLGHKKIIENELEGFGIRLNSKPPNI 179
Query 121 TIHKKDKGGIT 131
KKDKGGI
Sbjct 180 GFKKKDKGGIN 190
> CE24022
Length=366
Score = 215 bits (547), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 104/131 (79%), Positives = 114/131 (87%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+KTGDAR+G VGFPSVGKSTLL L G FSEVAAYEFTTLT VPGV YKGAK+QLLDLP
Sbjct 58 AKTGDARIGFVGFPSVGKSTLLCNLAGVFSEVAAYEFTTLTTVPGVIRYKGAKIQLLDLP 117
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDGKGRG+QVI VARTCSLIL+VLDV+ PL HK ++E ELEGFGIRLNKQPPNI
Sbjct 118 GIIEGAKDGKGRGKQVIAVARTCSLILMVLDVMKPLQHKKLLEYELEGFGIRLNKQPPNI 177
Query 121 TIHKKDKGGIT 131
+K+KGGI
Sbjct 178 GYKRKEKGGIN 188
> ECU05g0410i
Length=362
Score = 213 bits (541), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 102/131 (77%), Positives = 116/131 (88%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+KTG ARVG VGFPSVGKSTL++KLTGTFS VA+YEFTTLT VPGV Y GAK+Q+LDLP
Sbjct 58 AKTGIARVGFVGFPSVGKSTLMSKLTGTFSAVASYEFTTLTTVPGVLNYNGAKIQILDLP 117
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDGKGRG+QV+ VARTCSLI++ LDVL P++HK +IERELEG GIRLNKQPP I
Sbjct 118 GIIEGAKDGKGRGKQVLAVARTCSLIIVCLDVLKPVSHKAVIERELEGVGIRLNKQPPKI 177
Query 121 TIHKKDKGGIT 131
I KKDKGG+T
Sbjct 178 KITKKDKGGVT 188
> 7303536
Length=368
Score = 211 bits (538), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 102/131 (77%), Positives = 112/131 (85%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+KTGDARVG VGFPSVGKSTLL+ L G +SEVAAYEFTTLT VPG YKGAK+QLLDLP
Sbjct 60 AKTGDARVGFVGFPSVGKSTLLSNLAGVYSEVAAYEFTTLTTVPGCIKYKGAKIQLLDLP 119
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDGKGRGRQVI VARTC+LI +VLD L PL HK ++E ELEGFGIRLNK+PPNI
Sbjct 120 GIIEGAKDGKGRGRQVIAVARTCNLIFMVLDCLKPLGHKKLLEHELEGFGIRLNKKPPNI 179
Query 121 TIHKKDKGGIT 131
+KDKGGI
Sbjct 180 YYKRKDKGGIN 190
> SPAC9.07c
Length=366
Score = 201 bits (512), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 96/130 (73%), Positives = 111/130 (85%), Gaps = 0/130 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
++TG VG +GFPSVGKSTL+ +LTGT SE AAYEFTTLT VPGV Y GAK+Q+LDLP
Sbjct 59 ARTGIGTVGFIGFPSVGKSTLMTQLTGTRSEAAAYEFTTLTTVPGVLQYNGAKIQILDLP 118
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGAKDG+GRG+QVI VARTC+LI IVLDVL P++HK IIE ELEGFGIRLNK+PPNI
Sbjct 119 GIIEGAKDGRGRGKQVITVARTCNLIFIVLDVLKPMSHKRIIEEELEGFGIRLNKEPPNI 178
Query 121 TIHKKDKGGI 130
KK++GGI
Sbjct 179 VFKKKERGGI 188
> YAL036c
Length=369
Score = 197 bits (502), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 96/131 (73%), Positives = 111/131 (84%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
++TG A VG VGFPSVGKSTLL+KLTGT SE A YEFTTL VPGV YKGAK+Q+LDLP
Sbjct 61 ARTGVASVGFVGFPSVGKSTLLSKLTGTESEAAEYEFTTLVTVPGVIRYKGAKIQMLDLP 120
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GII+GAKDG+GRG+QVI VARTC+L+ I+LDV PL HK IIE+ELEG GIRLNK PP+I
Sbjct 121 GIIDGAKDGRGRGKQVIAVARTCNLLFIILDVNKPLHHKQIIEKELEGVGIRLNKTPPDI 180
Query 121 TIHKKDKGGIT 131
I KK+KGGI+
Sbjct 181 LIKKKEKGGIS 191
> 7300535
Length=363
Score = 180 bits (456), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 85/130 (65%), Positives = 102/130 (78%), Gaps = 0/130 (0%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPG 61
K+GDARV L+GFPSVGKST+L+ LT T SE A YEFTTLTC+PGV Y+GA +QLLDLPG
Sbjct 58 KSGDARVALIGFPSVGKSTMLSTLTKTESEAANYEFTTLTCIPGVIEYQGANIQLLDLPG 117
Query 62 IIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNIT 121
IIEGA GKGRGRQVI VART L+L++LD P H+ ++ERELE GIRLNK+ PNI
Sbjct 118 IIEGAAQGKGRGRQVIAVARTADLVLMMLDATKPNVHRELLERELESVGIRLNKRKPNIY 177
Query 122 IHKKDKGGIT 131
+K GG++
Sbjct 178 FKQKKGGGLS 187
> CE00039_1
Length=408
Score = 179 bits (455), Expect = 9e-46, Method: Composition-based stats.
Identities = 85/129 (65%), Positives = 100/129 (77%), Gaps = 0/129 (0%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPG 61
K+GDARV +VGFPSVGKSTLL+ +T T SE A YEFTTLTC+PGV Y GA +QLLDLPG
Sbjct 59 KSGDARVAMVGFPSVGKSTLLSSMTSTHSEAAGYEFTTLTCIPGVISYNGANIQLLDLPG 118
Query 62 IIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNIT 121
IIEGA GKGRGRQVI VA+T LIL++LD K ++ERELE GIRLNK+PPNI
Sbjct 119 IIEGASQGKGRGRQVISVAKTADLILMMLDAGKSDQQKMLLERELEAVGIRLNKKPPNIY 178
Query 122 IHKKDKGGI 130
+ +K GG+
Sbjct 179 VKQKKVGGV 187
> At1g72660
Length=399
Score = 172 bits (435), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 82/131 (62%), Positives = 99/131 (75%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+K G RV L+GFPSVGKSTLL LTGT SE A+YEFTTLTC+PGV +Y K+QLLDLP
Sbjct 58 TKYGHGRVALIGFPSVGKSTLLTMLTGTHSEAASYEFTTLTCIPGVIHYNDTKIQLLDLP 117
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGA +GKGRGRQVI VA++ L+L+VLD H+ I+ +ELE G+RLNK+PP I
Sbjct 118 GIIEGASEGKGRGRQVIAVAKSSDLVLMVLDASKSEGHRQILTKELEAVGLRLNKRPPQI 177
Query 121 TIHKKDKGGIT 131
KK GGI+
Sbjct 178 YFKKKKTGGIS 188
> Hs4557537
Length=364
Score = 171 bits (433), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 82/130 (63%), Positives = 99/130 (76%), Gaps = 0/130 (0%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPG 61
K+GDARV L+GFPSVGKST L+ +T T SE A+YEFTTLTC+PGV YKGA +QLLDLPG
Sbjct 59 KSGDARVALIGFPSVGKSTFLSLMTSTASEAASYEFTTLTCIPGVIEYKGANIQLLDLPG 118
Query 62 IIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNIT 121
IIEGA GKGRGRQVI VART +I+++LD ++++E+ELE GIRLNK PNI
Sbjct 119 IIEGAAQGKGRGRQVIAVARTADVIIMMLDATKGEVQRSLLEKELESVGIRLNKHKPNIY 178
Query 122 IHKKDKGGIT 131
K GGI+
Sbjct 179 FKPKKGGGIS 188
> At1g17470
Length=399
Score = 171 bits (432), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 82/131 (62%), Positives = 98/131 (74%), Gaps = 0/131 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+K G RV L+GFPSVGKSTLL LTGT SE A+YEFTTLTC+PGV +Y K+QLLDLP
Sbjct 58 TKYGHGRVALIGFPSVGKSTLLTMLTGTHSEAASYEFTTLTCIPGVIHYNDTKIQLLDLP 117
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GIIEGA +GKGRGRQVI VA++ L+L+VLD H+ I+ +ELE G+RLNK PP I
Sbjct 118 GIIEGASEGKGRGRQVIAVAKSSDLVLMVLDASKSEGHRQILTKELEAVGLRLNKTPPQI 177
Query 121 TIHKKDKGGIT 131
KK GGI+
Sbjct 178 YFKKKKTGGIS 188
> YGR173w
Length=368
Score = 160 bits (404), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 77/130 (59%), Positives = 98/130 (75%), Gaps = 0/130 (0%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
+K+GDARV L+G+PSVGKS+LL K+T T SE+A Y FTTLT VPGV Y+GA++Q++DLP
Sbjct 59 AKSGDARVVLIGYPSVGKSSLLGKITTTKSEIAHYAFTTLTSVPGVLKYQGAEIQIVDLP 118
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNI 120
GII GA GKGRGRQV+ ART L+L+VLD + +E+ELE GIRLNK+ PNI
Sbjct 119 GIIYGASQGKGRGRQVVATARTADLVLMVLDATKSEHQRASLEKELENVGIRLNKEKPNI 178
Query 121 TIHKKDKGGI 130
KK+ GG+
Sbjct 179 YYKKKETGGV 188
> SPBC354.01
Length=346
Score = 159 bits (402), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 78/129 (60%), Positives = 94/129 (72%), Gaps = 1/129 (0%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPG 61
K+GDARV +GFPSVGKSTLL+ +T T S A+YEFTTLT +PGV Y GA++Q+LDLPG
Sbjct 59 KSGDARVAFIGFPSVGKSTLLSAITKTKSATASYEFTTLTAIPGVLEYDGAEIQMLDLPG 118
Query 62 IIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPNIT 121
IIEGA G+G GRQ + ART LIL+VLD + IE ELE GIRLN+QPPN+T
Sbjct 119 IIEGASQGRG-GRQAVSAARTADLILMVLDATKAADQREKIEYELEQVGIRLNRQPPNVT 177
Query 122 IHKKDKGGI 130
+ K GGI
Sbjct 178 LTIKKNGGI 186
> ECU08g1270i
Length=362
Score = 139 bits (351), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 70/132 (53%), Positives = 92/132 (69%), Gaps = 5/132 (3%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLP 60
SK+GDARV L+G PSVGKSTLL+K+T T S+ A +EFTTL C+ G + +Q+LDLP
Sbjct 57 SKSGDARVVLIGLPSVGKSTLLSKITSTHSKAAEHEFTTLECISGKMHLNDTWIQVLDLP 116
Query 61 GIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKN--IIERELEGFGIRLNKQPP 118
GI+ GA +GRGRQVI +ART LIL+VLD P H++ I+E EL GIRLNK+ P
Sbjct 117 GIVSGASQNRGRGRQVISIARTADLILMVLD---PRRHEDRRILEDELHEMGIRLNKRKP 173
Query 119 NITIHKKDKGGI 130
++++ GI
Sbjct 174 DVSLTCTTSNGI 185
> 7297396
Length=381
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 53/124 (42%), Positives = 69/124 (55%), Gaps = 3/124 (2%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPG-VFYYKGAKVQLLDLPGIIE 64
A VGL+G+P+ GKSTLLN LT +VA Y FTTL G V Y ++ + DLPG++
Sbjct 206 ADVGLIGYPNAGKSTLLNALTRAKPKVAPYAFTTLRPHLGTVQYDDHVQLTIADLPGLVP 265
Query 65 GAKDGKGRGRQVIGVARTCSLILIVLDVL--NPLTHKNIIERELEGFGIRLNKQPPNITI 122
A KG G Q + A C+L+L VLD P TH + EL FG RL +P +
Sbjct 266 DAHRNKGLGIQFLKHAERCTLLLFVLDASAPEPWTHYEQLMHELRQFGGRLASRPQLVVA 325
Query 123 HKKD 126
+K D
Sbjct 326 NKLD 329
> Hs20560324
Length=406
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 44/124 (35%), Positives = 69/124 (55%), Gaps = 3/124 (2%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKG-AKVQLLDLPGIIE 64
A G+VGFP+ GKS+LL ++ VA+Y FTTL G+ +Y+G ++ + D+PGII
Sbjct 225 AHAGMVGFPNAGKSSLLRAISNARPAVASYPFTTLKPHVGIVHYEGHLQIAVADIPGIIR 284
Query 65 GAKDGKGRGRQVIGVARTCSLILIVLDVLN--PLTHKNIIERELEGFGIRLNKQPPNITI 122
GA +G G + C +L V+D+ P T + ++ ELE + L+ +P I
Sbjct 285 GAHQNRGLGSAFLRHIERCRFLLFVVDLSQPEPWTQVDDLKYELEMYEKGLSARPHAIVA 344
Query 123 HKKD 126
+K D
Sbjct 345 NKID 348
> SPAP8A3.11c
Length=419
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 57/98 (58%), Gaps = 11/98 (11%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTL--------TCVPGVFYYKGAKVQLL 57
+GLVG P+ GKSTLLN LT + S+V YEFTT+ T +P + + +L
Sbjct 237 CEIGLVGLPNAGKSTLLNCLTASKSKVGEYEFTTIYPKIGTIKTTMPDD--HSSFQYRLA 294
Query 58 DLPGIIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNP 95
D+PGII+GA DGKG G + ++ +V+D+ NP
Sbjct 295 DIPGIIKGASDGKGLGYDFLRHVERAKMLCLVIDI-NP 331
> CE12276
Length=358
Score = 67.4 bits (163), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 68/127 (53%), Gaps = 3/127 (2%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKG-AKVQLLDLPGIIE 64
A GLVGFP+ GKS+LL ++ +VA+Y FTTL GV +Y+ ++ + D+PG+IE
Sbjct 189 ATAGLVGFPNAGKSSLLRAISRAKPKVASYPFTTLHPHIGVVFYEDFEQIAVADIPGLIE 248
Query 65 GAKDGKGRGRQVIGVARTCSLILIVLDVLN-PLTHK-NIIERELEGFGIRLNKQPPNITI 122
+ KG G + C + VLD LT + ++ ELEG+ L + I I
Sbjct 249 DSHLNKGLGISFLKHIERCESLWYVLDYSTGSLTDQYKMLRVELEGYQKGLGDRASTIVI 308
Query 123 HKKDKGG 129
+K D G
Sbjct 309 NKIDLSG 315
> Hs22049574
Length=387
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 69/133 (51%), Gaps = 12/133 (9%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPG-VFYYKGAKVQLLDLPGIIE 64
A VGLVGFP+ GKS+LL+ ++ +A Y FTTL G + Y ++ + DLPG+IE
Sbjct 149 ADVGLVGFPNAGKSSLLSCVSHAKPAIADYAFTTLKPELGKIMYSDFKQISVADLPGLIE 208
Query 65 GAKDGKGRGRQVIGVARTCSLILIVLDV----LNPLTHKN-------IIERELEGFGIRL 113
GA KG G + + +L V+D+ L+ T ++ +ELE + L
Sbjct 209 GAHMNKGMGHKFLKHIERTRQLLFVVDISGFQLSSHTQYRTAFETIILLTKELELYKEEL 268
Query 114 NKQPPNITIHKKD 126
+P + ++K D
Sbjct 269 QTKPALLAVNKMD 281
> At5g18570
Length=699
Score = 64.7 bits (156), Expect = 5e-11, Method: Composition-based stats.
Identities = 45/125 (36%), Positives = 65/125 (52%), Gaps = 5/125 (4%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGV--FYYKGAKVQLLDLPGII 63
A VG+VG P+ GKSTLL+ ++ +A Y FTTL GV F Y V + DLPG++
Sbjct 379 ADVGIVGAPNAGKSTLLSVISAAQPTIANYPFTTLLPNLGVVSFDYDSTMV-VADLPGLL 437
Query 64 EGAKDGKGRGRQVIGVARTCSLILIVLD--VLNPLTHKNIIERELEGFGIRLNKQPPNIT 121
EGA G G G + + CS ++ V+D P + ELE F + ++P +
Sbjct 438 EGAHRGFGLGHEFLRHTERCSALVHVVDGSAPQPELEFEAVRLELELFSPEIAEKPYVVA 497
Query 122 IHKKD 126
+K D
Sbjct 498 YNKMD 502
> CE27734
Length=390
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 73/143 (51%), Gaps = 26/143 (18%)
Query 8 VGLVGFPSVGKSTLLNKLTGTFS-EVAAYEFTTLTCVPGVFYYKGAK----------VQL 56
+GL+GFP+ GKSTLL L S ++A Y FTT+ P V ++K + + +
Sbjct 164 IGLLGFPNAGKSTLLKALVPEKSVKIADYAFTTVN--PQVAFFKNKEEFSVEDPSFTLSI 221
Query 57 LDLPGIIEGAKDGKGRGRQVIGVARTCSLILIVLDVL-------------NPLTHKNIIE 103
DLPGIIEGA +G+G + + ++++V+D NPL ++
Sbjct 222 ADLPGIIEGASMNRGKGYKFLKHLEYADIVVMVVDCQGFQLKNELDCPFRNPLESIALLN 281
Query 104 RELEGFGIRLNKQPPNITIHKKD 126
RE+E + +L ++P ++K D
Sbjct 282 REVELYNQKLARKPVICVLNKID 304
> YHR168w
Length=499
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 36/105 (34%), Positives = 58/105 (55%), Gaps = 4/105 (3%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYK-GAKV-QLLDLPGII 63
A +GL+G P+ GKST+LNK++ ++ ++FTTL+ G G V + D+PGII
Sbjct 340 ADLGLIGLPNAGKSTILNKISNAKPKIGHWQFTTLSPTIGTVSLGFGQDVFTVADIPGII 399
Query 64 EGAKDGKGRGRQVIGVARTCSLILIVLDV--LNPLTHKNIIEREL 106
+GA KG G + + + + VLD+ NPL ++ E+
Sbjct 400 QGASLDKGMGLEFLRHIERSNGWVFVLDLSNKNPLNDLQLLIEEV 444
> At1g07620_2
Length=469
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 47/91 (51%), Gaps = 0/91 (0%)
Query 6 ARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEG 65
A VGLVG P+ GKSTLL L+ V Y FTTL G Y + + D+PG+I+G
Sbjct 280 ADVGLVGMPNAGKSTLLGALSRAKPRVGHYAFTTLRPNLGNVNYDDFSMTVADIPGLIKG 339
Query 66 AKDGKGRGRQVIGVARTCSLILIVLDVLNPL 96
A +G G + ++ V+D+ + L
Sbjct 340 AHQNRGLGHNFLRHIERTKVLAYVVDLASGL 370
> SPBC651.01c
Length=340
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 46/83 (55%), Gaps = 2/83 (2%)
Query 12 GFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEGAKDGKG 71
G+P+VGKS+ +NK+T +V Y FTT + G F YK + Q++D PGI++ +
Sbjct 175 GYPNVGKSSFMNKVTRAQVDVQPYAFTTKSLFVGHFDYKYLRWQVIDTPGILDHPLEQMN 234
Query 72 --RGRQVIGVARTCSLILIVLDV 92
+ + +A S +L +D+
Sbjct 235 TIEMQSITAMAHLRSAVLYFMDL 257
> SPBC428.15
Length=409
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 52/114 (45%), Gaps = 26/114 (22%)
Query 5 DARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTT-----------LTCVPGVF------ 47
D +G VG PS GKST+LN LT ++ + FTT + C F
Sbjct 4 DILIGFVGKPSSGKSTMLNALTDATAKTGNFPFTTIEPNRAIGYAQIECACSRFGLQDKC 63
Query 48 --YYKGAK-------VQLLDLPGIIEGAKDGKGRGRQVIGVARTCSLILIVLDV 92
Y G K +QLLD+ G+I GA GKG G + + R ++ V+DV
Sbjct 64 KPIYGGCKNGVRSIPIQLLDVAGLIPGAHAGKGLGNKFLDDLRHADALVHVVDV 117
> YGR210c
Length=411
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 35/114 (30%), Positives = 52/114 (45%), Gaps = 26/114 (22%)
Query 5 DARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLT----------------------C 42
D +G+VG PS GKST LN LT + V A+ FTT+ C
Sbjct 4 DPLIGIVGKPSSGKSTTLNSLTDAGAAVGAFPFTTIEPNQATGYLQVECACSRFGKEDLC 63
Query 43 VPGVFYYKGAK----VQLLDLPGIIEGAKDGKGRGRQVIGVARTCSLILIVLDV 92
P + K ++LLD+ G++ GA G+G G + + R ++ V+DV
Sbjct 64 KPNYGWCSKGKRHIPIKLLDVAGLVPGAHSGRGLGNKFLDDLRHADALIHVVDV 117
> Hs6912532
Length=633
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 54/102 (52%), Gaps = 4/102 (3%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEGAKDG 69
L G+P+VGKS+ +NK+T +V Y FTT + G YK + Q++D PGI++ +
Sbjct 173 LCGYPNVGKSSFINKVTRADVDVQPYAFTTKSLFVGHMDYKYLRWQVVDTPGILDHPLED 232
Query 70 KG--RGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGF 109
+ + + +A + +L V+D+ H + +LE F
Sbjct 233 RNTIEMQAITALAHLRAAVLYVMDLSEQCGHG--LREQLELF 272
> Hs14737772
Length=634
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 54/102 (52%), Gaps = 4/102 (3%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEGAKDG 69
L G+P+VGKS+ +NK+T +V Y FTT + G YK + Q++D PGI++ +
Sbjct 173 LCGYPNVGKSSFINKVTRADVDVQPYAFTTKSLFVGHMDYKYLRWQVVDTPGILDHPLED 232
Query 70 KG--RGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGF 109
+ + + +A + +L V+D+ H + +LE F
Sbjct 233 RNTIEMQAITALAHLRAAVLYVMDLSEQCGHG--LREQLELF 272
> 7303900
Length=652
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIE 64
+ GFP+VGKS+ +NK+T EV Y FTT + G YK + Q++D PGI++
Sbjct 173 ICGFPNVGKSSFINKITRADVEVQPYAFTTKSLYVGHTDYKYLRWQVIDTPGILD 227
> CE17218
Length=681
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 46/85 (54%), Gaps = 2/85 (2%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEGAKDG 69
L GFP+VGKS+ +N +T EV Y FTT G Y+ + Q++D PGI++ +
Sbjct 174 LCGFPNVGKSSFINNVTRADVEVQPYAFTTKALYVGHLDYRFLRWQVIDTPGILDQPLED 233
Query 70 KG--RGRQVIGVARTCSLILIVLDV 92
+ + V +A + +L ++DV
Sbjct 234 RNTIEMQAVTALAHLKASVLFMMDV 258
> ECU05g0800
Length=528
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 23/53 (43%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 12 GFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIE 64
GFP+VGKS+ + K++ EV Y FTT + G F YK + Q++D PGI++
Sbjct 174 GFPNVGKSSFVRKISRADVEVQPYPFTTKSLYVGHFDYKYLQWQVIDTPGILD 226
> At1g50920
Length=671
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 8 VGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIE 64
V + G+P+VGKS+ +NK+T +V Y FTT + G YK + Q++D PGI++
Sbjct 171 VLICGYPNVGKSSFMNKVTRADVDVQPYAFTTKSLFVGHTDYKYLRYQVIDTPGILD 227
> Hs18554526
Length=122
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCV 43
+TGDA +G V FPSVGKS L L G + E YEFT +T +
Sbjct 41 RTGDAYIGSVAFPSVGKSVDLRNLAGLYLEETPYEFTNMTAM 82
> 7291084
Length=399
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 19/100 (19%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTL--------TCVPG------VFYYKGA 52
R+G+VG P+VGKST N LT + + + F T+ VP V Y+K A
Sbjct 23 RIGIVGVPNVGKSTFFNVLTQSAAPAENFPFCTIKPNENKRRVPVPDERFDYLVEYHKPA 82
Query 53 KV-----QLLDLPGIIEGAKDGKGRGRQVIGVARTCSLIL 87
V ++D+ G+++GA +G+G G + C I
Sbjct 83 SVVPAYLNVVDIAGLVKGAAEGQGLGNDFLSHISACDAIF 122
> At1g56050
Length=418
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 19/113 (16%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLL--------- 57
+ G+VG P+VGKSTL N + ++ A + F T+ G+ +++Q+L
Sbjct 55 KAGIVGLPNVGKSTLFNAVENGKAQAANFPFCTIEPNVGIVAVPDSRLQVLSKLSGSQKT 114
Query 58 --------DLPGIIEGAKDGKGRGRQVIGVARTCSLILIVLDVL--NPLTHKN 100
D+ G+++GA G+G G + + R IL V+ N + H N
Sbjct 115 VPASIEFVDIAGLVKGASQGEGLGNKFLSHIREVDSILQVVRCFEDNDIIHVN 167
> YPL093w
Length=647
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIE 64
+ G+P+VGKS+ L +T + +V Y FTT + G F YK + Q +D PGI++
Sbjct 172 ICGYPNVGKSSFLRCITKSDVDVQPYAFTTKSLYVGHFDYKYLRFQAIDTPGILD 226
> At1g10300
Length=687
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIE 64
+ G P+VGKS+ +NK+T V Y FTT + G YK + Q++D PG+++
Sbjct 202 ICGCPNVGKSSFMNKVTRADVAVQPYAFTTKSLFLGHTDYKCLRYQVIDTPGLLD 256
> CE14708
Length=395
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 45/100 (45%), Gaps = 21/100 (21%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPG--------------VFYYKGA 52
+VG++G P+VGKST N LT SE A F T P V +YK A
Sbjct 23 KVGILGLPNVGKSTFFNVLTK--SEAQAENFPFCTIDPNESRVAVQDDRFDWLVNHYKPA 80
Query 53 K-----VQLLDLPGIIEGAKDGKGRGRQVIGVARTCSLIL 87
+ + D+ G+++GA +G+G G + C +
Sbjct 81 SKVPAFLNVTDIAGLVKGASEGQGLGNAFLSHVSACDALF 120
> Hs9506611
Length=582
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 52/103 (50%), Gaps = 20/103 (19%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVF-----YYKGAKVQLLDLPG 61
RVG+VG P+VGKS+L+N L + + ++ VPG+ Y ++LLD PG
Sbjct 254 RVGVVGLPNVGKSSLINSLKRSRA-------CSVGAVPGITKFMQEVYLDKFIRLLDAPG 306
Query 62 IIEGAKDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIER 104
I+ G +V + R C + + D + P+ + I++R
Sbjct 307 IVPGPNS------EVGTILRNCVHVQKLADPVTPV--ETILQR 341
> Hs9558757
Length=396
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 47/98 (47%), Gaps = 17/98 (17%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTL------TCVPG------VFYYKGAK- 53
++G+VG P+VGKST N LT + + + F T+ VP Y+K A
Sbjct 24 KIGIVGLPNVGKSTFFNVLTNSQASAENFPFCTIDPNESRVPVPDERFDFLCQYHKPASK 83
Query 54 ----VQLLDLPGIIEGAKDGKGRGRQVIGVARTCSLIL 87
+ ++D+ G+++GA +G+G G + C I
Sbjct 84 IPAFLNVVDIAGLVKGAHNGQGLGNAFLSHISACDGIF 121
> ECU06g1190
Length=369
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 55/135 (40%), Gaps = 29/135 (21%)
Query 1 SKTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLL--- 57
SK+ + +G+VG P+VGKSTL N LT Y F T+ G + +++ L
Sbjct 12 SKSNNLSMGIVGLPNVGKSTLFNFLTRNNVPAENYPFCTIDPSEGRVEIQDERIEFLAKK 71
Query 58 --------------DLPGIIEGAKDGKGRGRQVIGVARTCSLILIVL------------D 91
D+ G+++GA G G G + R I V+ D
Sbjct 72 YSPQNVARAYLSVTDIAGLVKGASAGVGLGNHFLDNIRNVDGIFHVVRCFEDTEVAHFED 131
Query 92 VLNPLTHKNIIEREL 106
++P+ I+ EL
Sbjct 132 GVDPIRDIEIVNEEL 146
> At1g30580
Length=394
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/129 (27%), Positives = 56/129 (43%), Gaps = 29/129 (22%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTL------TCVPGVFY------YK---- 50
++G+VG P+VGKSTL N LT + F T+ +P + YK
Sbjct 26 KIGIVGLPNVGKSTLFNTLTKLSIPAENFPFCTIEPNEARVNIPDERFDWLCQTYKPKSE 85
Query 51 -GAKVQLLDLPGIIEGAKDGKGRGRQVIGVARTCSLILIVL------------DVLNPLT 97
A +++ D+ G++ GA +G+G G + R I VL D+++P+
Sbjct 86 IPAFLEIHDIAGLVRGAHEGQGLGNNFLSHIRAVDGIFHVLRAFEDADIIHVDDIVDPVR 145
Query 98 HKNIIEREL 106
I EL
Sbjct 146 DLETITEEL 154
> At5g66470
Length=353
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 60/123 (48%), Gaps = 7/123 (5%)
Query 8 VGLVGFPSVGKSTLLNKLTGT-FSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGIIEGA 66
V +VG P+VGKSTL N++ G S V TT + G+ ++ L D PG+IE
Sbjct 58 VAVVGMPNVGKSTLSNQMIGQKISIVTDKPQTTRHRILGICSSPEYQMILYDTPGVIEKK 117
Query 67 --KDGKGRGRQVIGVARTCSLILIVLDVLNPLTHKNIIERELEGFGIRLNKQPPN-ITIH 123
+ + V A ++I++D T NI E EG G L K+PP + ++
Sbjct 118 MHRLDTMMMKNVRDAAINADCVVILVDACK--TPTNIEEVLKEGLG-DLEKKPPMLLVMN 174
Query 124 KKD 126
KKD
Sbjct 175 KKD 177
> Hs22064448
Length=437
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVP-GVFYYKGAKVQLLDLPGIIEG 65
RV L+G P+ GKSTL N+L G + + T C GV K +V LLD PGII
Sbjct 115 RVVLLGAPNAGKSTLSNQLLGRKVFPVSRKVHTTRCQALGVITEKETQVILLDTPGIISP 174
Query 66 AKDGK 70
K +
Sbjct 175 GKQKR 179
> Hs20555662
Length=607
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 12/67 (17%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPG-----VFYYKGAKVQL 56
K G +G VGFP+VGKS+L+N L G + +++ PG Y+ V+L
Sbjct 357 KDGVVTIGCVGFPNVGKSSLINGLVGR-------KVVSVSRTPGHTRYFQTYFLTPSVKL 409
Query 57 LDLPGII 63
D PG+I
Sbjct 410 CDCPGLI 416
> Hs4885289
Length=430
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 12/67 (17%)
Query 2 KTGDARVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPG-----VFYYKGAKVQL 56
K G +G VGFP+VGKS+L+N L G + +++ PG Y+ V+L
Sbjct 180 KDGVVTIGCVGFPNVGKSSLINGLVGR-------KVVSVSRTPGHTRYFQTYFLTPSVKL 232
Query 57 LDLPGII 63
D PG+I
Sbjct 233 CDCPGLI 239
> At1g80770
Length=451
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 10 LVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYKGAKVQLLDLPGII 63
LVG P+VGKS+L+ L+ E+ Y FTT + G + Q+ D PG++
Sbjct 258 LVGAPNVGKSSLVRILSTGKPEICNYPFTTRGILMGHIVLNYQRFQVTDTPGLL 311
> At1g30960
Length=437
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 44/91 (48%), Gaps = 6/91 (6%)
Query 8 VGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTC-VPGVFYYKGAKVQLLDLPGII--- 63
VG++G P+ GKS+L N + GT A+ + T T V GV +V D PG++
Sbjct 156 VGIIGPPNAGKSSLTNFMVGTKVAAASRKTNTTTHEVLGVLTKGDTQVCFFDTPGLMLKK 215
Query 64 --EGAKDGKGRGRQVIGVARTCSLILIVLDV 92
G KD K R + +++++ DV
Sbjct 216 SGYGYKDIKARVQNAWTSVDLFDVLIVMFDV 246
> 7300220
Length=581
Score = 37.4 bits (85), Expect = 0.008, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 12/62 (19%)
Query 7 RVGLVGFPSVGKSTLLNKLTGTFSEVAAYEFTTLTCVPGVFYYK-----GAKVQLLDLPG 61
RVG+VG P+VGKS+++N LT S + PGV +K++L+D PG
Sbjct 269 RVGVVGIPNVGKSSIINSLTRGRS-------CMVGSTPGVTKSMQEVELDSKIKLIDCPG 321
Query 62 II 63
I+
Sbjct 322 IV 323
Lambda K H
0.319 0.142 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40