bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3084_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
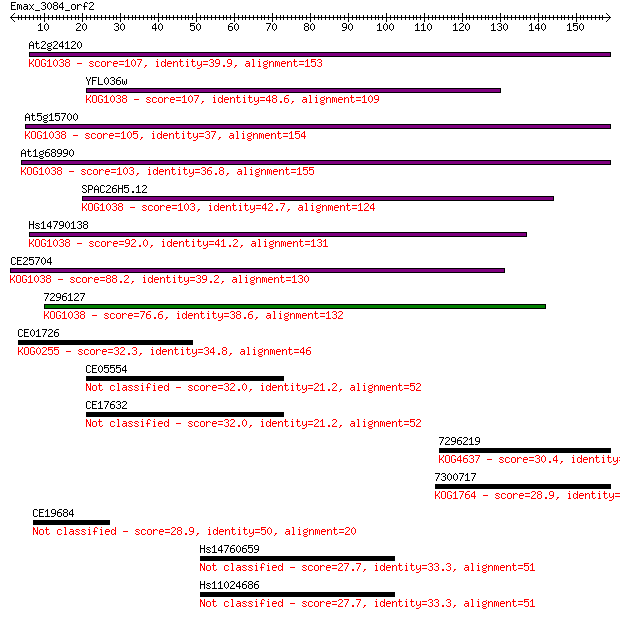
At2g24120 107 7e-24
YFL036w 107 9e-24
At5g15700 105 3e-23
At1g68990 103 9e-23
SPAC26H5.12 103 2e-22
Hs14790138 92.0 5e-19
CE25704 88.2 6e-18
7296127 76.6 2e-14
CE01726 32.3 0.41
CE05554 32.0 0.54
CE17632 32.0 0.56
7296219 30.4 1.3
7300717 28.9 4.6
CE19684 28.9 4.8
Hs14760659 27.7 9.3
Hs11024686 27.7 9.5
> At2g24120
Length=993
Score = 107 bits (268), Expect = 7e-24, Method: Composition-based stats.
Identities = 61/153 (39%), Positives = 83/153 (54%), Gaps = 6/153 (3%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRS 65
WL + + + PV W P +GL QPY ++ IR++LQ L G++ V
Sbjct 832 WLGDCAKIIASDNHPVRWITP-LGLPVVQPYCRSERHLIRTSLQVLALQREGNTVDVR-- 888
Query 66 KQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQFV 125
KQR FPPNF+HSLD HMM+TA+ C ++ A VHDSYW HA +DT+ LR++FV
Sbjct 889 KQRTAFPPNFVHSLDGTHMMMTAVAC-REAGLNFAGVHDSYWTHACDVDTMNRILREKFV 947
Query 126 ALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
LY L+ L S + P PP P +G
Sbjct 948 ELYNTPILEDLLQSFQESYPNLV--FPPVPKRG 978
> YFL036w
Length=1351
Score = 107 bits (267), Expect = 9e-24, Method: Composition-based stats.
Identities = 53/111 (47%), Positives = 75/111 (67%), Gaps = 6/111 (5%)
Query 21 VAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA--PVSRSKQRLGFPPNFIHS 78
V W P +GL QPYR ++Q+ + LQ T+ +S A PV+ +Q+ G PPNFIHS
Sbjct 1108 VIWTTP-LGLPIVQPYREESKKQVETNLQ--TVFISDPFAVNPVNARRQKAGLPPNFIHS 1164
Query 79 LDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQFVALYQ 129
LDA+HM+L+A +C G + +D A+VHDSYW HA IDT+ LR+QF+ L++
Sbjct 1165 LDASHMLLSAAEC-GKQGLDFASVHDSYWTHASDIDTMNVVLREQFIKLHE 1214
> At5g15700
Length=1011
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 57/154 (37%), Positives = 82/154 (53%), Gaps = 6/154 (3%)
Query 5 KWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSR 64
+W + + + V W P +GL QPY + + ++++LQ +L D V R
Sbjct 849 RWFGECAKIIASENETVRWTTP-LGLPVVQPYHQMGTKLVKTSLQTLSLQHETDQVIVRR 907
Query 65 SKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQF 124
QR FPPNFIHSLD +HMM+TA+ C + A VHDS+W HA +D L LR++F
Sbjct 908 --QRTAFPPNFIHSLDGSHMMMTAVAC-KRAGVCFAGVHDSFWTHACDVDKLNIILREKF 964
Query 125 VALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
V LY L++L +S + P PP P +G
Sbjct 965 VELYSQPILENLLESFEQSFPH--LDFPPLPERG 996
> At1g68990
Length=976
Score = 103 bits (258), Expect = 9e-23, Method: Composition-based stats.
Identities = 57/155 (36%), Positives = 83/155 (53%), Gaps = 6/155 (3%)
Query 4 KKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVS 63
K W + + + V W P +GL QPYR + +++ LQ TL S ++ V
Sbjct 813 KSWFGDCAKIIASENNAVCWTTP-LGLPVVQPYRKPGRHLVKTTLQVLTL--SRETDKVM 869
Query 64 RSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
+Q F PNFIHSLD +HMM+TA+ C + A VHDS+W HA +D + T LR++
Sbjct 870 ARRQMTAFAPNFIHSLDGSHMMMTAVAC-NRAGLSFAGVHDSFWTHACDVDVMNTILREK 928
Query 124 FVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
FV LY+ L++L +S + P PP P +G
Sbjct 929 FVELYEKPILENLLESFQKSFPD--ISFPPLPERG 961
> SPAC26H5.12
Length=1120
Score = 103 bits (257), Expect = 2e-22, Method: Composition-based stats.
Identities = 53/124 (42%), Positives = 73/124 (58%), Gaps = 2/124 (1%)
Query 20 PVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRSKQRLGFPPNFIHSL 79
PV W + L QPYR+ + +QIR+ LQ + +A V KQ FPPNFIHSL
Sbjct 945 PVVWTT-LLNLPIVQPYRNYKSRQIRTNLQTVFIEERDRTATVQPHKQATAFPPNFIHSL 1003
Query 80 DAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQFVALYQHNPLQSLYDS 139
DA HM +T L C ++I+ AAVHDSYW HA +D + + LR+ FV L+ +N ++ L
Sbjct 1004 DATHMFMTCLKC-SEQNINFAAVHDSYWTHACDVDQMNSLLREAFVLLHSNNIMERLKQE 1062
Query 140 VKLR 143
+ R
Sbjct 1063 FEER 1066
> Hs14790138
Length=1230
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 74/133 (55%), Gaps = 4/133 (3%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYR-SLQQQQIRSALQRHTLHVSGD-SAPVS 63
WL + + + G V W P +G+ QPYR + +QI +Q T +GD S +
Sbjct 1053 WLTESARLISHMGSVVEWVTP-LGVPVIQPYRLDSKVKQIGGGIQSITYTHNGDISRKPN 1111
Query 64 RSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
KQ+ GFPPNFIHSLD++HMMLTAL C + + +VHD YW HA + + R+Q
Sbjct 1112 TRKQKNGFPPNFIHSLDSSHMMLTALHCY-RKGLTFVSVHDCYWTHAADVSVMNQVCREQ 1170
Query 124 FVALYQHNPLQSL 136
FV L+ LQ L
Sbjct 1171 FVRLHSEPILQDL 1183
> CE25704
Length=1154
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 51/130 (39%), Positives = 68/130 (52%), Gaps = 14/130 (10%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
M K W ++ + V W P +GL QPY L +++ + L A
Sbjct 999 MALKDWFRLIAKGSSDLMKTVEWITP-LGLPVVQPYCKLVERKGKLIL-----------A 1046
Query 61 PVSRSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATAL 120
PV KQ FPPNF+HSLD+ HMMLT+L+C R I AAVHD +W HA +D +
Sbjct 1047 PVPM-KQVDAFPPNFVHSLDSTHMMLTSLNC-AQRGITFAAVHDCFWTHANSVDQMNEIC 1104
Query 121 RQQFVALYQH 130
RQQFV+L+
Sbjct 1105 RQQFVSLHSQ 1114
> 7296127
Length=1369
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 51/138 (36%), Positives = 75/138 (54%), Gaps = 16/138 (11%)
Query 10 VSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRS---- 65
+S VC+ V W P +GL QPY +Q+++ + R VS + P+
Sbjct 1192 ISGVCSQN---VEWVTP-LGLPVVQPY---NRQEMKHS-PRSGFKVSAN-MPMDLYERPN 1242
Query 66 --KQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
KQ+ FPPNFIHSLD++HMMLT+L C + I +VHD +W HA + L R+Q
Sbjct 1243 ILKQKNAFPPNFIHSLDSSHMMLTSLHC-ERQGITFVSVHDCFWTHANTVPELNRMCREQ 1301
Query 124 FVALYQHNPLQSLYDSVK 141
FVAL+ L+ L + ++
Sbjct 1302 FVALHSQPILEQLSEFMR 1319
> CE01726
Length=541
Score = 32.3 bits (72), Expect = 0.41, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query 3 TKKWLDAVSTVCTAAGV---PVAWKVPAVGLLCEQPYRSLQQQQIRSAL 48
T++W D S VC A P+ WK+P + EQ +R + + R L
Sbjct 213 TRRW-DTASYVCAAISALIFPILWKLPESPVFLEQKHRIEEANEAREQL 260
> CE05554
Length=432
Score = 32.0 bits (71), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 11/52 (21%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 21 VAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRSKQRLGFP 72
+ W++P +G +P+++ + + L T+++ G ++ +KQ G P
Sbjct 1 MIWEIPIIGDFITKPFKAAKLYEFYDRLHLFTVYLLGFFVLLTGAKQHFGNP 52
> CE17632
Length=434
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 11/52 (21%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 21 VAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRSKQRLGFP 72
+ W++P +G +P+++ + + L T+++ G ++ +KQ G P
Sbjct 1 MIWEIPIIGDFITKPFKAAKLYEFYDRLHLFTVYLLGFFVLLTGAKQHFGNP 52
> 7296219
Length=496
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 21/49 (42%), Positives = 27/49 (55%), Gaps = 6/49 (12%)
Query 114 DTLATALRQQFVALYQHNPLQ----SLYDSVKLRLPQQAAKLPPPPAKG 158
DTL T LR V +++NPLQ L + + L +QAA L PPP G
Sbjct 410 DTLTTTLRWP-VLYWKNNPLQVQMIQLQEEMDLEY-EQAATLRPPPMMG 456
> 7300717
Length=634
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 113 IDTLATALRQQFVALYQHNPLQSLYDSVKLRLPQQAAKLPP-PPAKG 158
+DT + L Q + L P SLYD++K+ + + +LP PA G
Sbjct 243 LDTWRSVLHNQVMPLVSIGPDASLYDAIKILIHSRIHRLPVIDPATG 289
> CE19684
Length=1341
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 10/20 (50%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 7 LDAVSTVCTAAGVPVAWKVP 26
L+ + VC AAG+P+ WK+P
Sbjct 942 LENMPAVCEAAGLPMYWKMP 961
> Hs14760659
Length=706
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 51 HTLHVSGDSAPVSRSKQRLG-FPPNFIHSLDAAHMMLTALDCIGTRHIDMAA 101
HT V DS RSK+ L + +HS+ A + T L+ + +H D+A+
Sbjct 208 HTYPVGEDSLVSDRSKKELSPVLTSEVHSVRAGRHLATKLNILVQQHFDLAS 259
> Hs11024686
Length=706
Score = 27.7 bits (60), Expect = 9.5, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 51 HTLHVSGDSAPVSRSKQRLG-FPPNFIHSLDAAHMMLTALDCIGTRHIDMAA 101
HT V DS RSK+ L + +HS+ A + T L+ + +H D+A+
Sbjct 208 HTYPVGEDSLVSDRSKKELSPVLTSEVHSVRAGRHLATKLNILVQQHFDLAS 259
Lambda K H
0.322 0.132 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40