bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3030_orf1
Length=183
Score E
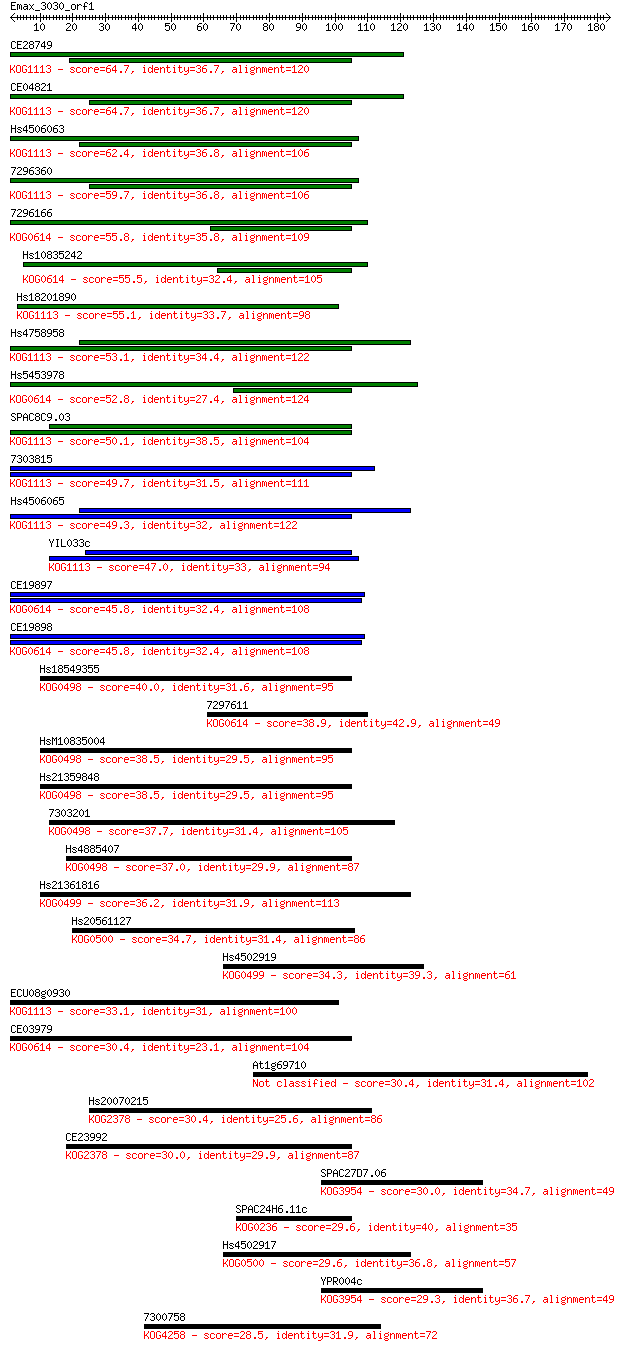
Sequences producing significant alignments: (Bits) Value
CE28749 64.7 9e-11
CE04821 64.7 1e-10
Hs4506063 62.4 5e-10
7296360 59.7 3e-09
7296166 55.8 5e-08
Hs10835242 55.5 6e-08
Hs18201890 55.1 7e-08
Hs4758958 53.1 3e-07
Hs5453978 52.8 4e-07
SPAC8C9.03 50.1 3e-06
7303815 49.7 3e-06
Hs4506065 49.3 4e-06
YIL033c 47.0 2e-05
CE19897 45.8 4e-05
CE19898 45.8 5e-05
Hs18549355 40.0 0.003
7297611 38.9 0.006
HsM10835004 38.5 0.008
Hs21359848 38.5 0.008
7303201 37.7 0.013
Hs4885407 37.0 0.022
Hs21361816 36.2 0.040
Hs20561127 34.7 0.11
Hs4502919 34.3 0.14
ECU08g0930 33.1 0.33
CE03979 30.4 1.8
At1g69710 30.4 1.9
Hs20070215 30.4 2.1
CE23992 30.0 2.7
SPAC27D7.06 30.0 2.9
SPAC24H6.11c 29.6 3.6
Hs4502917 29.6 3.9
YPR004c 29.3 4.2
7300758 28.5 7.5
> CE28749
Length=334
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 61/121 (50%), Gaps = 2/121 (1%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQDGGGFLE 59
++ DL + E VA A + F G V + G GD F I +GEA + QK D F
Sbjct 208 ILADLDQWERANVADALERCDFEPGTHVVEQGQPGDEFFIILEGEANVLQKRSDDAPFDV 267
Query 60 PARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTY 119
G +YFGE+AL+ R ATVVA T L LD+ FE ++ P++E + + S Y
Sbjct 268 VGHLGMSDYFGEIALLLDRPRAATVVAKTHLKCIKLDRNRFERVMGPVRE-ILKRDVSNY 326
Query 120 K 120
Sbjct 327 N 327
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 47/86 (54%), Gaps = 6/86 (6%)
Query 19 VERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIKST 78
VE+ A G+T+ + G GD F I KG + ++ +G FGE+ALI T
Sbjct 109 VEKSA-GETIIEQGEEGDNFYVIDKGTVDVYVNHE---YVLTINEGGS--FGELALIYGT 162
Query 79 RRTATVVAGTELVLFCLDKESFETLL 104
R ATV+A T++ L+ +D+ ++ +L
Sbjct 163 PRAATVIAKTDVKLWAIDRLTYRRIL 188
> CE04821
Length=376
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 61/121 (50%), Gaps = 2/121 (1%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQDGGGFLE 59
++ DL + E VA A + F G V + G GD F I +GEA + QK D F
Sbjct 250 ILADLDQWERANVADALERCDFEPGTHVVEQGQPGDEFFIILEGEANVLQKRSDDAPFDV 309
Query 60 PARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTY 119
G +YFGE+AL+ R ATVVA T L LD+ FE ++ P++E + + S Y
Sbjct 310 VGHLGMSDYFGEIALLLDRPRAATVVAKTHLKCIKLDRNRFERVMGPVRE-ILKRDVSNY 368
Query 120 K 120
Sbjct 369 N 369
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 5/80 (6%)
Query 25 GQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIKSTRRTATV 84
G+T+ + G GD F I KG + ++ +G FGE+ALI T R ATV
Sbjct 156 GETIIEQGEEGDNFYVIDKGTVDVYVNHE---YVLTINEGGS--FGELALIYGTPRAATV 210
Query 85 VAGTELVLFCLDKESFETLL 104
+A T++ L+ +D+ ++ +L
Sbjct 211 IAKTDVKLWAIDRLTYRRIL 230
> Hs4506063
Length=381
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 57/107 (53%), Gaps = 1/107 (0%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQDGGGFLE 59
++ L + E VA A + +F +GQ + G GD F I +G A + Q+ + F+E
Sbjct 255 ILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEGSAAVLQRRSENEEFVE 314
Query 60 PARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSP 106
R G +YFGE+AL+ + R ATVVA L LD+ FE +L P
Sbjct 315 VGRLGPSDYFGEIALLMNRPRAATVVARGPLKCVKLDRPRFERVLGP 361
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 44/83 (53%), Gaps = 5/83 (6%)
Query 22 FAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIKSTRRT 81
F G+TV + G GD F I +GE + + G+G FGE+ALI T R
Sbjct 158 FIAGETVIQQGDEGDNFYVIDQGETDVYVNNEWATSV-----GEGGSFGELALIYGTPRA 212
Query 82 ATVVAGTELVLFCLDKESFETLL 104
ATV A T + L+ +D++S+ +L
Sbjct 213 ATVKAKTNVKLWGIDRDSYRRIL 235
> 7296360
Length=299
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 56/108 (51%), Gaps = 2/108 (1%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAIT--QKPQDGGGFL 58
++ L + E VA + + F +G+T+ K G GD F I +G A+ Q+ + G
Sbjct 170 ILESLDKWERLTVADSLETCSFDDGETIVKQGAAGDDFYIILEGCAVVLQQRSEQGEDPA 229
Query 59 EPARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSP 106
E R G +YFGE+AL+ R ATVVA L LD+ FE +L P
Sbjct 230 EVGRLGSSDYFGEIALLLDRPRAATVVARGPLKCVKLDRARFERVLGP 277
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 41/87 (47%), Gaps = 19/87 (21%)
Query 25 GQTVFKMGFHGDMFCFIYKGEA-------ITQKPQDGGGFLEPARQGKGEYFGEMALIKS 77
G+ + + G GD F I GE + +GG F GE+ALI
Sbjct 76 GENIIQQGDEGDNFYVIDVGEVDVFVNSELVTTISEGGSF------------GELALIYG 123
Query 78 TRRTATVVAGTELVLFCLDKESFETLL 104
T R ATV A T++ L+ +D++S+ +L
Sbjct 124 TPRAATVRAKTDVKLWGIDRDSYRRIL 150
> 7296166
Length=768
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 62/116 (53%), Gaps = 11/116 (9%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQK-----PQDG 54
L+ +L EE + +A ++E +A G + + G GD F I +G +TQK P++
Sbjct 304 LLMNLSEELLAKIADVLELEFYAAGTYIIRQGTAGDSFFLISQGNVRVTQKLTPTSPEET 363
Query 55 GGFLEPARQGKGEYFGEMALIKSTRRTATVVAGTELV-LFCLDKESFETLLSPLKE 109
E +G+YFGE ALI +RTA ++A + V LD++SF+ L+ L E
Sbjct 364 ----ELRTLSRGDYFGEQALINEDKRTANIIALSPGVECLTLDRDSFKRLIGDLCE 415
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Query 62 RQGKGEYFGEMALIKSTRRTATVVAGTELV-LFCLDKESFETLL 104
+ G G+ FGE+A++ + RTA++ +E ++ LD+ F+ ++
Sbjct 241 KMGAGKAFGELAILYNCTRTASIRVLSEAARVWVLDRRVFQQIM 284
> Hs10835242
Length=686
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 53/108 (49%), Gaps = 5/108 (4%)
Query 5 LPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQK---PQDGGGFLEPA 61
LPEE + +A + + G+ + + G GD F I KG + P + FL
Sbjct 240 LPEEILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGTVNVTREDSPSEDPVFLRTL 299
Query 62 RQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSPLKE 109
GKG++FGE AL RTA V+A + +D++SF+ L+ L +
Sbjct 300 --GKGDWFGEKALQGEDVRTANVIAAEAVTCLVIDRDSFKHLIGGLDD 345
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 64 GKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
G G+ FGE+A++ + RTATV + L+ +D++ F+T++
Sbjct 176 GPGKVFGELAILYNCTRTATVKTLVNVKLWAIDRQCFQTIM 216
> Hs18201890
Length=572
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 33/101 (32%), Positives = 51/101 (50%), Gaps = 3/101 (2%)
Query 3 RDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDG-GGFLEPA 61
R+ E +A+ + ERF + + K G G+ F FIY G K +DG FL+P
Sbjct 118 RNYAEPLQLLLAKVMRFERFGRRRVIIKKGQKGNSFYFIYLGTVAITKDEDGSSAFLDPH 177
Query 62 RQ--GKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESF 100
+ KG FGEM ++ ++ R +T+V E +D+E F
Sbjct 178 PKLLHKGSCFGEMDVLHASVRRSTIVCMEETEFLVVDREDF 218
> Hs4758958
Length=404
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 56/108 (51%), Gaps = 8/108 (7%)
Query 22 FAEGQTVFKMGFHGDMFCFIYKGE------AITQKPQDGGGF-LEPARQGKGEYFGEMAL 74
+ +G+ + G D F I GE + T+ +DGG +E AR KG+YFGE+AL
Sbjct 282 YKDGERIITQGEKADSFYIIESGEVSILIRSRTKSNKDGGNQEVEIARCHKGQYFGELAL 341
Query 75 IKSTRRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTYKRQ 122
+ + R A+ A ++ +D ++FE LL P + M + S Y+ Q
Sbjct 342 VTNKPRAASAYAVGDVKCLVMDVQAFERLLGPCMDIM-KRNISHYEEQ 388
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 54/110 (49%), Gaps = 13/110 (11%)
Query 1 LVRDLPEEEIYAVARACKVERFAEG-QTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLE 59
L ++L +E++ V A ER + + V G GD F I +G +D
Sbjct 139 LFKNLDQEQLSQVLDAM-FERIVKADEHVIDQGDDGDNFYVIERGTYDILVTKDN----- 192
Query 60 PARQGKGEY-----FGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
+ G+Y FGE+AL+ +T R AT+VA +E L+ LD+ +F ++
Sbjct 193 -QTRSVGQYDNRGSFGELALMYNTPRAATIVATSEGSLWGLDRVTFRRII 241
> Hs5453978
Length=762
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 63/128 (49%), Gaps = 4/128 (3%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
L+++LPE+++ + +VE + +G + + G G F + KG+ + +G +
Sbjct 286 LLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKGKVKVTQSTEGHDQPQL 345
Query 61 ARQ-GKGEYFGEMALIKSTRRTATVVA-GTELVLFCLDKESFETLLSPLKE--KMAAKAA 116
+ KGEYFGE ALI R+A ++A ++ +D+E+F + +E K
Sbjct 346 IKTLQKGEYFGEKALISDDVRSANIIAEENDVACLVIDRETFNQTVGTFEELQKYLEGYV 405
Query 117 STYKRQDE 124
+ R DE
Sbjct 406 ANLNRDDE 413
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 69 FGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
FGE+A++ + RTA+V A T + + LD+E F+ ++
Sbjct 231 FGELAILYNCTRTASVKAITNVKTWALDREVFQNIM 266
> SPAC8C9.03
Length=412
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 44/92 (47%), Gaps = 4/92 (4%)
Query 13 VARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEM 72
+A A + + G V + G G+ F I GEA K G KG+YFGE+
Sbjct 290 IADALQTVVYQAGSIVIRQGDIGNQFYLIEDGEAEVVKNGKGVV----VTLTKGDYFGEL 345
Query 73 ALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
ALI T R ATV A T L L DK +F LL
Sbjct 346 ALIHETVRNATVQAKTRLKLATFDKPTFNRLL 377
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 57/118 (48%), Gaps = 16/118 (13%)
Query 1 LVRDLPEEEIYAVARACKVERFAE-GQTVFKMGFHGDMFCFIYKGE-AITQKPQDG---- 54
L ++L EE V A +R E G V G GD F + +GE + ++P+
Sbjct 143 LFKNLDEEHYNEVLNAMTEKRIGEAGVAVIVQGAVGDYFYIVEQGEFDVYKRPELNITPE 202
Query 55 -------GGFLEPARQGKGEYFGEMALIKSTRRTATVVAGT-ELVLFCLDKESFETLL 104
G ++ GEYFGE+AL+ + R A+VV+ T V++ LD+ SF ++
Sbjct 203 EVLSSGYGNYITTI--SPGEYFGELALMYNAPRAASVVSKTPNNVIYALDRTSFRRIV 258
> 7303815
Length=411
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 58/112 (51%), Gaps = 2/112 (1%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
+++ L E +A A + + G+ + K G D FI +G + QD +E
Sbjct 242 MLKALQNYERMNLADALVSKSYDNGERIIKQGDAADGMYFIEEGTVSVRMDQDDAE-VEI 300
Query 61 ARQGKGEYFGEMALIKSTRRTATVVA-GTELVLFCLDKESFETLLSPLKEKM 111
++ GKG+YFGE+AL+ R A+V A G + L LD E+FE ++ L + +
Sbjct 301 SQLGKGQYFGELALVTHRPRAASVYATGGVVKLAFLDTEAFERIMGFLTDVL 352
Score = 37.0 bits (84), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 46/104 (44%), Gaps = 5/104 (4%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
L R L +E++ V A + G + + G GD F I G K +
Sbjct 124 LFRSLEKEQMNQVLDAMFERKVQPGDFIIRQGDDGDNFYVIESG---VYKVYINDKHINT 180
Query 61 ARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
FGE+AL+ + R ATV A T +L+ +D+++F +L
Sbjct 181 YNHTG--LFGELALLYNMPRAATVQAETSGLLWAMDRQTFRRIL 222
> Hs4506065
Length=418
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 53/107 (49%), Gaps = 7/107 (6%)
Query 22 FAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQ-----DGGGFLEPARQGKGEYFGEMALI 75
+ +G+ + G D F + GE IT K + + G +E R +G+YFGE+AL+
Sbjct 297 YNDGEQIIAQGDSADSFFIVESGEVKITMKRKGKSEVEENGAVEMPRCSRGQYFGELALV 356
Query 76 KSTRRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTYKRQ 122
+ R A+ A + +D ++FE LL P E M A TY+ Q
Sbjct 357 TNKPRAASAHAIGTVKCLAMDVQAFERLLGPCMEIMKRNIA-TYEEQ 402
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 50/104 (48%), Gaps = 1/104 (0%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
L ++L E++ V A + +G+ V G GD F I +G DG G
Sbjct 154 LFKNLDPEQMSQVLDAMFEKLVKDGEHVIDQGDDGDNFYVIDRGTFDIYVKCDGVGRCVG 213
Query 61 ARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
+G FGE+AL+ +T R AT+ A + L+ LD+ +F ++
Sbjct 214 NYDNRGS-FGELALMYNTPRAATITATSPGALWGLDRVTFRRII 256
> YIL033c
Length=416
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 5/81 (6%)
Query 24 EGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIKSTRRTAT 83
+G T+ K G GD F + KG + D + G G FGE+AL+ ++ R AT
Sbjct 207 KGATIIKQGDQGDYFYVVEKG-TVDFYVNDN----KVNSSGPGSSFGELALMYNSPRAAT 261
Query 84 VVAGTELVLFCLDKESFETLL 104
VVA ++ +L+ LD+ +F +L
Sbjct 262 VVATSDCLLWALDRLTFRKIL 282
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 43/94 (45%), Gaps = 4/94 (4%)
Query 13 VARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEM 72
+A A + + G+T+ + G G+ F I G K G G + + +YFGE+
Sbjct 314 LADALDTKIYQPGETIIREGDQGENFYLIEYGAVDVSK--KGQGVINKLKDH--DYFGEV 369
Query 73 ALIKSTRRTATVVAGTELVLFCLDKESFETLLSP 106
AL+ R ATV A + L K F+ LL P
Sbjct 370 ALLNDLPRQATVTATKRTKVATLGKSGFQRLLGP 403
> CE19897
Length=780
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 59/115 (51%), Gaps = 12/115 (10%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQDGGGFLE 59
+ ++L E+ I +A + + G + + G GD F I G+ +TQ+ + G E
Sbjct 319 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQVKVTQQIE---GETE 375
Query 60 PAR---QGKGEYFGEMALIKSTRRTATVVA---GTELVLFCLDKESFETLLSPLK 108
P +G++FGE AL+ RTA ++A G E++ LD+ESF L+ L+
Sbjct 376 PREIRVLNQGDFFGERALLGEEVRTANIIAQAPGVEVL--TLDRESFGKLIGDLE 428
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 50/107 (46%), Gaps = 5/107 (4%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
++ L +E+I + R GQ V + G GD + +GE + G L
Sbjct 201 FLKQLAKEQIIELVNCMYEMRARAGQWVIQEGEPGDRLFVVAEGELQVSRE---GALLGK 257
Query 61 ARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSPL 107
R G GE+A++ + RTA+V A T++ L+ LD+ F+ + L
Sbjct 258 MR--AGTVMGELAILYNCTRTASVQALTDVQLWVLDRSVFQMITQRL 302
> CE19898
Length=737
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 59/115 (51%), Gaps = 12/115 (10%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEA-ITQKPQDGGGFLE 59
+ ++L E+ I +A + + G + + G GD F I G+ +TQ+ + G E
Sbjct 276 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQVKVTQQIE---GETE 332
Query 60 PAR---QGKGEYFGEMALIKSTRRTATVVA---GTELVLFCLDKESFETLLSPLK 108
P +G++FGE AL+ RTA ++A G E++ LD+ESF L+ L+
Sbjct 333 PREIRVLNQGDFFGERALLGEEVRTANIIAQAPGVEVL--TLDRESFGKLIGDLE 385
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 50/107 (46%), Gaps = 5/107 (4%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
++ L +E+I + R GQ V + G GD + +GE + G L
Sbjct 158 FLKQLAKEQIIELVNCMYEMRARAGQWVIQEGEPGDRLFVVAEGELQVSRE---GALLGK 214
Query 61 ARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLLSPL 107
R G GE+A++ + RTA+V A T++ L+ LD+ F+ + L
Sbjct 215 MR--AGTVMGELAILYNCTRTASVQALTDVQLWVLDRSVFQMITQRL 259
> Hs18549355
Length=675
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 42/97 (43%), Gaps = 9/97 (9%)
Query 10 IYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKG--EAITQKPQDGGGFLEPARQGKGE 67
+ AV + E F G V + G G FI G + + +D R G
Sbjct 338 VTAVLTKLRFEVFQPGDLVVREGSVGRKMYFIQHGLLSVLARGARD-------TRLTDGS 390
Query 68 YFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
YFGE+ L+ RRTA+V A T L+ L + F +L
Sbjct 391 YFGEICLLTRGRRTASVRADTYCRLYSLSVDHFNAVL 427
> 7297611
Length=1003
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 35/52 (67%), Gaps = 5/52 (9%)
Query 61 ARQGKGEYFGEMALIKSTRRTATVVA---GTELVLFCLDKESFETLLSPLKE 109
A + +G+YFGE AL+ + R A+V A GTE+++ LD+E+F + L +K+
Sbjct 610 ANRKRGDYFGEQALLNADVRQASVYADAPGTEVLM--LDREAFISYLGTIKQ 659
> HsM10835004
Length=889
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 9/97 (9%)
Query 10 IYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKG--EAITQKPQDGGGFLEPARQGKGE 67
+ A+ K E F G + + G G FI G +T+ ++ + G
Sbjct 553 VTAMLTKLKFEVFQPGDYIIREGTIGKKMYFIQHGVVSVLTKGNKE-------MKLSDGS 605
Query 68 YFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
YFGE+ L+ RRTA+V A T L+ L ++F +L
Sbjct 606 YFGEICLLTRGRRTASVRADTYCRLYSLSVDNFNEVL 642
> Hs21359848
Length=889
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 9/97 (9%)
Query 10 IYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKG--EAITQKPQDGGGFLEPARQGKGE 67
+ A+ K E F G + + G G FI G +T+ ++ + G
Sbjct 553 VTAMLTKLKFEVFQPGDYIIREGTIGKKMYFIQHGVVSVLTKGNKE-------MKLSDGS 605
Query 68 YFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
YFGE+ L+ RRTA+V A T L+ L ++F +L
Sbjct 606 YFGEICLLTRGRRTASVRADTYCRLYSLSVDNFNEVL 642
> 7303201
Length=1307
Score = 37.7 bits (86), Expect = 0.013, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 43/107 (40%), Gaps = 6/107 (5%)
Query 13 VARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEM 72
V K E F G + K G G FI +G + G G YFGE+
Sbjct 1116 VVTKLKYEVFQPGDIIIKEGTIGTKMYFIQEG--VVDIVMANGEV--ATSLSDGSYFGEI 1171
Query 73 ALIKSTRRTATVVAGTELVLFCLDKESFETLLS--PLKEKMAAKAAS 117
L+ + RR A+V A T LF L + F +L PL K A+
Sbjct 1172 CLLTNARRVASVRAETYCNLFSLSVDHFNCVLDQYPLMRKTMETVAA 1218
> Hs4885407
Length=1203
Score = 37.0 bits (84), Expect = 0.022, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 41/89 (46%), Gaps = 9/89 (10%)
Query 18 KVERFAEGQTVFKMGFHGDMFCFIYKG--EAITQKPQDGGGFLEPARQGKGEYFGEMALI 75
+ E F G + + G G FI G +T+ ++ + G YFGE+ L+
Sbjct 612 RFEVFQPGDYIIREGTIGKKMYFIQHGVVSVLTKGNKE-------TKLADGSYFGEICLL 664
Query 76 KSTRRTATVVAGTELVLFCLDKESFETLL 104
RRTA+V A T L+ L ++F +L
Sbjct 665 TRGRRTASVRADTYCRLYSLSVDNFNEVL 693
> Hs21361816
Length=809
Score = 36.2 bits (82), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 50/118 (42%), Gaps = 7/118 (5%)
Query 10 IYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYF 69
IY + K + G V K G G I GE DG L + G F
Sbjct 533 IYDMLLRLKSVLYLPGDFVCKKGEIGKEMYIIKHGEVQVLGGPDGTKVLVTLKAG--SVF 590
Query 70 GEMALIKS---TRRTATVVAGTELVLFCLDKESFETLLS--PLKEKMAAKAASTYKRQ 122
GE++L+ + RRTA VVA L LDK++ + +L P E++ K A +Q
Sbjct 591 GEISLLAAGGGNRRTANVVAHGFANLLTLDKKTLQEILVHYPDSERILMKKARVLLKQ 648
> Hs20561127
Length=575
Score = 34.7 bits (78), Expect = 0.11, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 41/92 (44%), Gaps = 9/92 (9%)
Query 20 ERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIK--- 76
+ ++ G+ V + G G I +G+ G + A G G YFGE+++I
Sbjct 367 QTYSPGEYVCRKGDIGQEMYIIREGQLAVVADD---GITQYAVLGAGLYFGEISIINIKG 423
Query 77 ---STRRTATVVAGTELVLFCLDKESFETLLS 105
RRTA + + LFCL KE +LS
Sbjct 424 NMSGNRRTANIKSLGYSDLFCLSKEDLREVLS 455
> Hs4502919
Length=1245
Score = 34.3 bits (77), Expect = 0.14, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 66 GEYFGEMALIK---STRRTATVVAGTELVLFCLDKESFETLLS--PLKEKMAAKAASTYK 120
G FGE++L+ RRTA VVA LF LDK+ +L P +K+ K A
Sbjct 1019 GSVFGEISLLAVGGGNRRTANVVAHGFTNLFILDKKDLNEILVHYPESQKLLRKKARRML 1078
Query 121 RQDEAP 126
R + P
Sbjct 1079 RSNNKP 1084
> ECU08g0930
Length=312
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 49/105 (46%), Gaps = 10/105 (9%)
Query 1 LVRDLP-----EEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGG 55
LV D+P E+ + + ++ G V G G + GE K GG
Sbjct 113 LVSDIPFGFLNPEQKTRLIESTELIEIKRGTFVMHEGEIGSQMYIVASGEFEVTK---GG 169
Query 56 GFLEPARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESF 100
L + KG +FGE+AL+ + RTATV A T+ ++ +++ SF
Sbjct 170 TLLR--KLTKGCFFGEIALLHNIPRTATVKAVTDGKVWVVEQTSF 212
> CE03979
Length=581
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 24/104 (23%), Positives = 44/104 (42%), Gaps = 5/104 (4%)
Query 1 LVRDLPEEEIYAVARACKVERFAEGQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEP 60
+R+L +I ++ A G + + G G + I +G+ K +E
Sbjct 36 FLRNLDATQIEKISSAMYPVEVPAGAIIIRQGDLGSIMYVIQEGKVQVVKDNRFVRTME- 94
Query 61 ARQGKGEYFGEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
G FGE+A++ RTATV A L+ +++ F ++
Sbjct 95 ----DGALFGELAILHHCERTATVRAIESCHLWAIERNVFHAIM 134
> At1g69710
Length=1028
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 54/114 (47%), Gaps = 18/114 (15%)
Query 75 IKST--RRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTYKRQDEAPLTASEM 132
+KST RR+ V A L K+SF ++ LKE++ A+ ++ ++E T ++
Sbjct 819 VKSTSPRRSYEVAAAESKQL----KDSFNQDMAGLKEQVEQLASKAHQLEEELEKTKRQL 874
Query 133 KSADGSALAPVLATRDAGEVC----------AKKRTRKNSLALLNYSKIRDQEK 176
K A R A EV A+K+++K+S++ SK D+EK
Sbjct 875 KVVTAMAADEAEENRSAKEVIRSLTTQLKEMAEKQSQKDSIS--TNSKHTDKEK 926
> Hs20070215
Length=881
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 5/87 (5%)
Query 25 GQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIKSTRRTATV 84
G +F G G + I+KG G G + +G + FG++AL+ R AT+
Sbjct 228 GTVLFSQGDKGTSWYIIWKGS--VNVVTHGKGLVTTLHEG--DDFGQLALVNDAPRAATI 283
Query 85 VAGTELVLFC-LDKESFETLLSPLKEK 110
+ + F +DK+ F ++ ++ K
Sbjct 284 ILREDNCHFLRVDKQDFNRIIKDVEAK 310
> CE23992
Length=1234
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 7/89 (7%)
Query 18 KVERFAE-GQTVFKMGFHGDMFCFIYKGEAITQKPQDGGGFLEPARQGKGEYFGEMALIK 76
KVE++ G VF+ G G + + KG + G + R+G + FG++AL+
Sbjct 588 KVEQYVHAGSVVFRQGEIGVYWYIVLKGAV---EVNVNGKIVCLLREG--DDFGKLALVN 642
Query 77 STRRTATVVAGTELVLF-CLDKESFETLL 104
R AT+V + +F +DK F +L
Sbjct 643 DLPRAATIVTYEDDSMFLVVDKHHFNQIL 671
> SPAC27D7.06
Length=341
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 96 DKESFETLLSPLKEKMAAKAASTYKRQDEAPLTASEMKSADGSALAPVL 144
DKE+FE +L+PL +K+ A +T D S G +AP L
Sbjct 237 DKETFERILTPLADKLGAAIGATRVAVDSGYADNSLQIGQTGKIIAPKL 285
> SPAC24H6.11c
Length=958
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 70 GEMALIKSTRRTATVVAGTELVLFCLDKESFETLL 104
GE+ T ATVVA + ++ LD+E +ET+L
Sbjct 887 GELPFFSKTCYNATVVAELDSAVWILDREGWETML 921
> Hs4502917
Length=694
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 29/63 (46%), Gaps = 8/63 (12%)
Query 66 GEYFGEMALIK------STRRTATVVAGTELVLFCLDKESFETLLSPLKEKMAAKAASTY 119
G YFGE++++ RRTA + + LFCL K+ L+ E A KA
Sbjct 544 GSYFGEISILNIKGSKSGNRRTANIRSIGYSDLFCLSKDDLMEALTEYPE--AKKALEEK 601
Query 120 KRQ 122
RQ
Sbjct 602 GRQ 604
> YPR004c
Length=344
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 96 DKESFETLLSPLKEKMAAKAASTYKRQDEAPLTASEMKSADGSALAPVL 144
DKE+FE LLSPL + + A +T D S G +AP L
Sbjct 236 DKETFEKLLSPLADVLHAAIGATRASVDNGLCDNSLQIGQTGKVVAPNL 284
> 7300758
Length=1742
Score = 28.5 bits (62), Expect = 7.5, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 34/82 (41%), Gaps = 10/82 (12%)
Query 42 YKGEAITQKP-----QDGGGFLEPARQGKGEYFGEMALIKSTRRT-----ATVVAGTELV 91
Y G+ IT P + EP GK + LI S R T++ GTE +
Sbjct 614 YNGQCITHCPTGYQKSENKRMCEPCPGGKCDKECSSGLIDSLERAREFHGCTIITGTEPL 673
Query 92 LFCLDKESFETLLSPLKEKMAA 113
+ +ES ++ LK +AA
Sbjct 674 TISIKRESGAHVMDELKYGLAA 695
Lambda K H
0.315 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40