bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3027_orf2
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
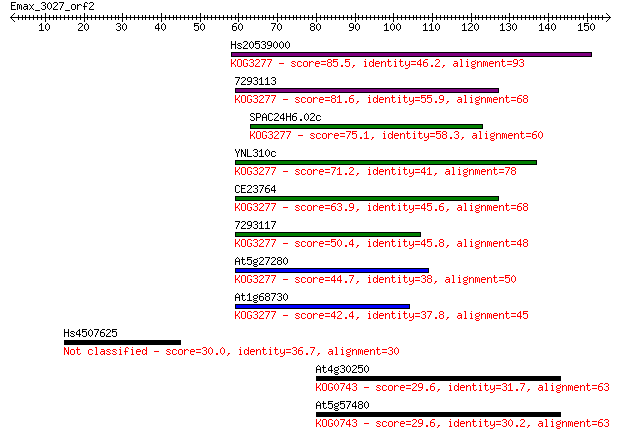
Hs20539000 85.5 3e-17
7293113 81.6 5e-16
SPAC24H6.02c 75.1 5e-14
YNL310c 71.2 8e-13
CE23764 63.9 1e-10
7293117 50.4 2e-06
At5g27280 44.7 7e-05
At1g68730 42.4 4e-04
Hs4507625 30.0 2.1
At4g30250 29.6 2.5
At5g57480 29.6 2.5
> Hs20539000
Length=178
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/96 (44%), Positives = 60/96 (62%), Gaps = 3/96 (3%)
Query 58 YLTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGE--EESDVET 115
+ LV+TC C RS+K+ SK+AY +GVVIV CPGC N H++AD L WF + + ++E
Sbjct 68 HYQLVYTCKVCGTRSSKRISKLAYHQGVVIVTCPGCQNHHIIADNLGWFSDLNGKRNIEE 127
Query 116 ILAAKGEKVLRTLSASQV-LDIEGLDCSSQTAAPSA 150
IL A+GE+V R + L +E + TAAP A
Sbjct 128 ILTARGEQVHRVAGEGALELVLEAAGAPTSTAAPEA 163
> 7293113
Length=83
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/70 (54%), Positives = 49/70 (70%), Gaps = 2/70 (2%)
Query 59 LTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGEEES--DVETI 116
+ L++TC C R+ K SK+AY RGVVIV C GCSN HL+AD L WF + + ++E I
Sbjct 1 MQLIYTCKVCQTRNMKTISKLAYQRGVVIVTCEGCSNHHLIADNLNWFTDLDGKRNIEEI 60
Query 117 LAAKGEKVLR 126
LA KGEKV+R
Sbjct 61 LAEKGEKVVR 70
> SPAC24H6.02c
Length=175
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/60 (58%), Positives = 39/60 (65%), Gaps = 0/60 (0%)
Query 63 FTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGEEESDVETILAAKGE 122
FTC CN RS FSK AY G V+V+CP C N HL+AD LK F EE +E ILA KGE
Sbjct 80 FTCTVCNTRSNHNFSKQAYHNGTVLVQCPKCKNRHLMADHLKIFSEERVTIEDILAKKGE 139
> YNL310c
Length=205
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 46/78 (58%), Gaps = 0/78 (0%)
Query 59 LTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGEEESDVETILA 118
+ + FTC KCN RS+ SK AY +G V++ CP C HL+AD LK F + VE ++
Sbjct 100 MMIAFTCKKCNTRSSHTMSKQAYEKGTVLISCPHCKVRHLIADHLKIFHDHHVTVEQLMK 159
Query 119 AKGEKVLRTLSASQVLDI 136
A GE+V + + + DI
Sbjct 160 ANGEQVSQDVGDLEFEDI 177
> CE23764
Length=119
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 44/70 (62%), Gaps = 2/70 (2%)
Query 59 LTLVFTCCKCNKRSA-KKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGE-EESDVETI 116
L+L +TC CN R K F+K +Y +GVVIV C GC N H++AD + WF + + ++E
Sbjct 35 LSLSYTCKVCNSREGPKTFAKSSYEKGVVIVTCSGCHNHHIIADNIGWFEDFKGKNIEDH 94
Query 117 LAAKGEKVLR 126
L +GE V R
Sbjct 95 LKTRGEAVKR 104
> 7293117
Length=191
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 59 LTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWF 106
+ LV+ C CN R+ K S+ AY GVVI++C GC+ HL+ D L F
Sbjct 102 MELVYRCKLCNTRNTKTISEEAYYSGVVILQCDGCAVDHLIKDNLGLF 149
> At5g27280
Length=212
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 59 LTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRLKWFGE 108
+ + FTC C +R+ + + AYT G V V+C GC+ H + D L F E
Sbjct 121 MRVAFTCNVCGQRTTRAINPHAYTDGTVFVQCCGCNVFHKLVDNLNLFHE 170
> At1g68730
Length=353
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 59 LTLVFTCCKCNKRSAKKFSKVAYTRGVVIVRCPGCSNLHLVADRL 103
L + FTC C +R+ + ++ AY +G+V V+C GC H + D L
Sbjct 106 LQVEFTCNSCGERTKRLINRHAYEKGLVFVQCAGCLKHHKLVDNL 150
> Hs4507625
Length=251
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 15 TRPAAATTAPPPVAEGPSVDFEGVHDVRFD 44
+RP PP + EG VDF+ +H R +
Sbjct 32 SRPVVPPLIPPKIPEGERVDFDDIHRKRME 61
> At4g30250
Length=512
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 3/66 (4%)
Query 80 AYTRGVVIVRCPGCSNLHLVADRLKWFGEEESDVE-TILAAKGE--KVLRTLSASQVLDI 136
A+ RG ++ PG L+A + G + D+E T + E K+L S+ ++ I
Sbjct 236 AWKRGYLLYGPPGTGKSSLIAAMANYLGYDIYDLELTEVQNNSELRKLLMKTSSKSIIVI 295
Query 137 EGLDCS 142
E +DCS
Sbjct 296 EDIDCS 301
> At5g57480
Length=520
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 34/66 (51%), Gaps = 3/66 (4%)
Query 80 AYTRGVVIVRCPGCSNLHLVADRLKWFGEEESDVE-TILAAKGE--KVLRTLSASQVLDI 136
A+ RG ++ PG ++A + G + D+E T + + E K+L S+ ++ I
Sbjct 235 AWKRGYLLYGPPGTGKSSMIAAMANYLGYDIYDLELTEVHSNSELRKLLMKTSSKSIIVI 294
Query 137 EGLDCS 142
E +DCS
Sbjct 295 EDIDCS 300
Lambda K H
0.316 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40