bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
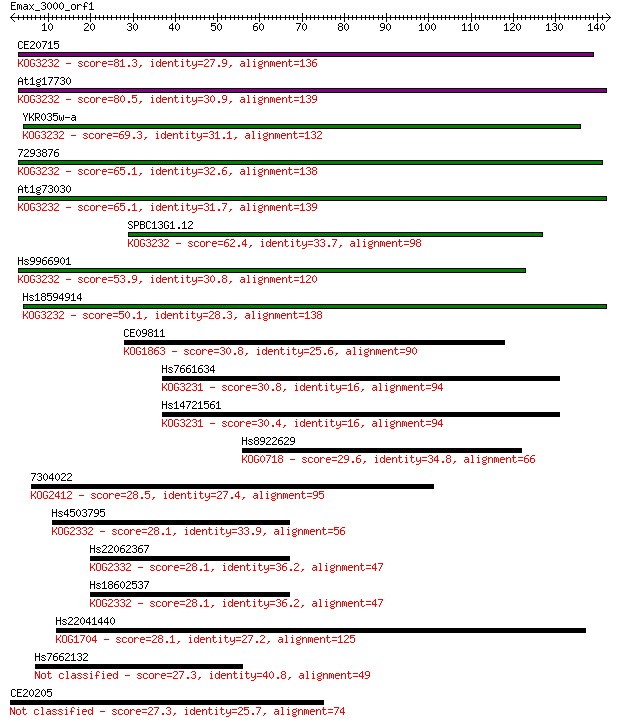
Query= Emax_3000_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
CE20715 81.3 5e-16
At1g17730 80.5 1e-15
YKR035w-a 69.3 2e-12
7293876 65.1 4e-11
At1g73030 65.1 4e-11
SPBC13G1.12 62.4 3e-10
Hs9966901 53.9 1e-07
Hs18594914 50.1 1e-06
CE09811 30.8 0.79
Hs7661634 30.8 0.96
Hs14721561 30.4 0.99
Hs8922629 29.6 2.1
7304022 28.5 4.6
Hs4503795 28.1 5.0
Hs22062367 28.1 5.1
Hs18602537 28.1 5.6
Hs22041440 28.1 5.8
Hs7662132 27.3 8.5
CE20205 27.3 8.7
> CE20715
Length=205
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/136 (27%), Positives = 81/136 (59%), Gaps = 0/136 (0%)
Query 3 DLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 62
DL A++L++ + RC ++ E+DK+ A+ + N+E A++ A+N +RKK E +N + M+
Sbjct 16 DLKFAAKQLEKNAQRCEKDEKVEKDKLTAAIKKGNKEVAQVHAENAIRKKNEAVNYIKMA 75
Query 63 SKLEAVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRS 122
++++AVA+R+ A ++T + V + A++ ++ + ++ F + F+DLDV +
Sbjct 76 ARIDAVAARVQTAATQKRVTASMSGVVKAMESAMKSMNLEKVQQLMDRFERDFEDLDVTT 135
Query 123 DSVSSLLDVSTSSALP 138
++ +D +T P
Sbjct 136 KTMEKTMDGTTVLNAP 151
> At1g17730
Length=203
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 88/144 (61%), Gaps = 10/144 (6%)
Query 3 DLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 62
+L ++ L+RQ+ +C +E E+ K+++A+ + N +GARI+A+N +RK+ E +N L +S
Sbjct 13 ELKFTSKSLQRQARKCEKEERSEKLKVKKAIEKGNMDGARIYAENAIRKRSEQMNYLRLS 72
Query 63 SKLEAVASRLDGAHRSHQLTTKIHSVASGL-----SGALRKLDSSSSLREIELFSKLFDD 117
S+L+AV +RLD + +T + ++ L +G L+K+ + ++ F K F +
Sbjct 73 SRLDAVVARLDTQAKMATITKSMTNIVKSLESSLTTGNLQKMSET-----MDSFEKQFVN 127
Query 118 LDVRSDSVSSLLDVSTSSALPSGK 141
++V+++ + + + STS + P G+
Sbjct 128 MEVQAEFMDNAMAGSTSLSTPEGE 151
> YKR035w-a
Length=204
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 80/132 (60%), Gaps = 4/132 (3%)
Query 4 LGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSS 63
L +++L++Q+++ +E QE +K++RAL +N++ +RI+A N +RKK E L L ++S
Sbjct 16 LKFTSKQLQKQANKASKEEKQETNKLKRAL-NENEDISRIYASNAIRKKNERLQLLKLAS 74
Query 64 KLEAVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSD 123
++++VASR+ A Q++ + V G+ AL+ ++ ++ F + F+DLD
Sbjct 75 RVDSVASRVQTAVTMRQVSASMGQVCKGMDKALQNMNLQQITMIMDKFEQQFEDLDT--- 131
Query 124 SVSSLLDVSTSS 135
SV+ D+ +S
Sbjct 132 SVNVYEDMGVNS 143
> 7293876
Length=198
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 45/140 (32%), Positives = 84/140 (60%), Gaps = 4/140 (2%)
Query 3 DLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 62
+L +EL+R S +C +E E+ K ++A+ + N + ARI A+N +R+K + +N L MS
Sbjct 7 NLKFAVKELERNSKKCEKEEKLEKAKAKKAIQKGNMDVARIHAENAIRQKNQAVNYLRMS 66
Query 63 SKLEAVASRLDGAHRSHQLTTKIHSVASGLSGALR--KLDSSSSLREIELFSKLFDDLDV 120
++++AVASR+ A + ++T + V + A++ L+ SSL +E F F+DLDV
Sbjct 67 ARVDAVASRVQSALTTRKVTGSMAGVVKAMDAAMKGMNLEKISSL--MEKFESQFEDLDV 124
Query 121 RSDSVSSLLDVSTSSALPSG 140
+S + + + ++++P G
Sbjct 125 QSSVMEGTMSDTVTTSVPQG 144
> At1g73030
Length=203
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 44/144 (30%), Positives = 88/144 (61%), Gaps = 10/144 (6%)
Query 3 DLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 62
DL ++ L+RQS +C +E E+ K+++A+ + N +GARI+A+N +RK+ E +N L ++
Sbjct 13 DLKFTSKSLQRQSRKCEKEEKAEKLKVKKAIEKGNMDGARIYAENAIRKRSEQMNYLRLA 72
Query 63 SKLEAVASRLDGAHRSHQLTTKIHSVASGL-----SGALRKLDSSSSLREIELFSKLFDD 117
S+L+AV +RLD + +T + ++ L +G L+K+ + ++ F K F +
Sbjct 73 SRLDAVVARLDTQAKMTTITKSMTNIVKSLESSLATGNLQKMSET-----MDSFEKQFVN 127
Query 118 LDVRSDSVSSLLDVSTSSALPSGK 141
++V+++ + + + STS + P G+
Sbjct 128 MEVQAEFMENAMAGSTSLSTPEGE 151
> SPBC13G1.12
Length=178
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 58/98 (59%), Gaps = 7/98 (7%)
Query 29 IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTKIHSV 88
I A+ + N E ARI+A N +RK+QE LN L +SS+++AV+SRL Q + +V
Sbjct 14 IAIAITKGNSEIARIYASNAIRKQQESLNLLKLSSRIDAVSSRL-------QTAVTMRAV 66
Query 89 ASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVS 126
+ ++G +R +D + +E+ S++ D + + D V+
Sbjct 67 SGNMAGVVRGMDRAMKTMNLEMISQVMDKFEAQFDDVN 104
> Hs9966901
Length=196
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 70/120 (58%), Gaps = 0/120 (0%)
Query 3 DLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 62
+L A+EL R + +C +E E+ KI++A+ + N E ARI A+N +R+K + +N L MS
Sbjct 7 NLKFAAKELSRSAKKCDKEEKAEKAKIKKAIQKGNMEVARIHAENAIRQKNQAVNFLRMS 66
Query 63 SKLEAVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRS 122
++++AVA+R+ A ++T + V + L+ ++ ++ F F+ LDV++
Sbjct 67 ARVDAVAARVQTAVTMGKVTKSMAGVVKSMDATLKTMNLEKISALMDKFEHQFETLDVQT 126
> Hs18594914
Length=417
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/152 (25%), Positives = 69/152 (45%), Gaps = 37/152 (24%)
Query 4 LGIKARELKRQSDRCYRESLQERDKIR--------------RALLRQNQEGARIFAQNYV 49
L A+EL R + +C +E E+ KI+ +A+ + N+E ARI A+N +
Sbjct 192 LKFAAKELNRNAKKCDKEEKAEKAKIKSNSESGNLSYEDSWKAIQKGNKEAARIHAENAI 251
Query 50 RKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIE 109
+K + ++ L MS+++ AVA R A V G + AL ++
Sbjct 252 HQKNQAIDFLRMSARVGAVAVRGQNA------------VTMGRTSAL-----------MD 288
Query 110 LFSKLFDDLDVRSDSVSSLLDVSTSSALPSGK 141
F F+ LD+++ + + ST+ P G+
Sbjct 289 KFKHQFETLDIQTQQMEDKMSSSTTLITPQGQ 320
> CE09811
Length=1230
Score = 30.8 bits (68), Expect = 0.79, Method: Composition-based stats.
Identities = 23/96 (23%), Positives = 45/96 (46%), Gaps = 6/96 (6%)
Query 28 KIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTT---- 83
++++ +QN E +++Q RK G N + S + + V+SR+ SH++ T
Sbjct 605 ELKKRTRQQNGEVKYVYSQRTPRKSHNGTNGTNSSPQKQPVSSRVAALLSSHEIPTCGHG 664
Query 84 --KIHSVASGLSGALRKLDSSSSLREIELFSKLFDD 117
I + G A+ + + + LRE + K+ D
Sbjct 665 KMSIDPILYGDVKAVSRAPAIALLREYDFRVKIVYD 700
> Hs7661634
Length=213
Score = 30.8 bits (68), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 15/94 (15%), Positives = 48/94 (51%), Gaps = 0/94 (0%)
Query 37 NQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTKIHSVASGLSGAL 96
N+E ++ A+ V +++ + +SSK+ +++++ + ++ + + A +
Sbjct 54 NKEACKVLAKQLVHLRKQKTRTFAVSSKVTSMSTQTKVMNSQMKMAGAMSTTAKTMQAVN 113
Query 97 RKLDSSSSLREIELFSKLFDDLDVRSDSVSSLLD 130
+K+D +L+ ++ F K +++ + ++ LD
Sbjct 114 KKMDPQKTLQTMQNFQKENMKMEMTEEMINDTLD 147
> Hs14721561
Length=213
Score = 30.4 bits (67), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 15/94 (15%), Positives = 48/94 (51%), Gaps = 0/94 (0%)
Query 37 NQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTKIHSVASGLSGAL 96
N+E ++ A+ V +++ + +SSK+ +++++ + ++ + + A +
Sbjct 54 NKEACKVLAKQLVHLRKQKTRTFAVSSKVTSMSTQTKVMNSQMKMAGAMSTTAKTMQAVN 113
Query 97 RKLDSSSSLREIELFSKLFDDLDVRSDSVSSLLD 130
+K+D +L+ ++ F K +++ + ++ LD
Sbjct 114 KKMDPQKTLQTMQNFQKENMKMEMTEEMINDTLD 147
> Hs8922629
Length=559
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 38/76 (50%), Gaps = 10/76 (13%)
Query 56 LNSLHMSSKLEAVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIE------ 109
+N +H+S +EA + D A S L+T+ + ++ ALR++ S+ E+E
Sbjct 159 INKMHISQSIEAPLTATDTAILSGSLSTQNGNGGGSINFALRRVTSAKGWGELEFGAGDL 218
Query 110 ---LFS-KLFDDLDVR 121
LF KLF +L R
Sbjct 219 QGPLFGLKLFRNLTPR 234
> 7304022
Length=677
Score = 28.5 bits (62), Expect = 4.6, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 49/105 (46%), Gaps = 10/105 (9%)
Query 6 IKARELKRQSDRCYRESLQ----ERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHM 61
I REL+ + RC R+ L+ ++D +R + Q ++ R Q + +E SL +
Sbjct 90 IVRRELEEEGKRCVRQQLKAIRDKQDAMRLSRETQQRKEERQRDQLQQKALRERNESLLI 149
Query 62 SSKLEAVASRLDGAHRS-----HQLTTKIHSVA-SGLSGALRKLD 100
+ A++L+ R Q+ K+H +A G+S R+ +
Sbjct 150 QKADQMTAAQLEAQQREQLALRQQIDQKLHKLALEGVSRCQRRFN 194
> Hs4503795
Length=190
Score = 28.1 bits (61), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 6/61 (9%)
Query 11 LKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKK-----QEGLNSLHMSSKL 65
LK + +S +ER+ + + QNQ G RIF Q+ ++K + GLN++ + L
Sbjct 49 LKNFAKYFLHQSHEEREHAEKLMKLQNQRGGRIFLQD-IKKPDCDDWESGLNAMECALHL 107
Query 66 E 66
E
Sbjct 108 E 108
> Hs22062367
Length=183
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 6/52 (11%)
Query 20 RESLQERDKIRRALLRQNQEGARIFAQNYVRKK-----QEGLNSLHMSSKLE 66
+S +ER+ + + QNQ G RIF Q+ ++K + GLN++ + LE
Sbjct 58 HQSHEEREHAEKLMKLQNQRGGRIFLQD-IKKPDCDDWESGLNAMECALHLE 108
> Hs18602537
Length=146
Score = 28.1 bits (61), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 6/52 (11%)
Query 20 RESLQERDKIRRALLRQNQEGARIFAQNYVRKK-----QEGLNSLHMSSKLE 66
+S +ER+ + + QNQ G RIF Q+ ++K + GLN++ + LE
Sbjct 21 HQSHEEREHAEKLMKLQNQGGGRIFLQD-IKKPDCDDWESGLNAMECALHLE 71
> Hs22041440
Length=1082
Score = 28.1 bits (61), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 57/130 (43%), Gaps = 19/130 (14%)
Query 12 KRQSDRCYRESLQERDKIRRALLR-----QNQEGARIFAQNYVRKKQEGLNSLHMSSKLE 66
+R+S + YRE +QE+++ R L ++QE A Q Y+ + +S E
Sbjct 405 RRKSIKTYREIVQEKERRERELHEAYKNARSQEEAEGILQQYIER-------FTIS---E 454
Query 67 AVASRLDGAHRSHQLTTKIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVS 126
AV RL+ ++ + HS LS L + LR+ L F + + +
Sbjct 455 AVLERLEMP----KILERSHSTEPNLSSFLNDPNPMKYLRQQSLPPPKFTATVETTIARA 510
Query 127 SLLDVSTSSA 136
S+LD S S+
Sbjct 511 SVLDTSMSAG 520
> Hs7662132
Length=1313
Score = 27.3 bits (59), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 32/52 (61%), Gaps = 5/52 (9%)
Query 7 KARELKRQSDRCYRESLQERDKIRRALLRQNQEGAR---IFAQNYVRKKQEG 55
+AR+L +Q D C RE+ + R ++R LL ++G R + AQ +R+ QEG
Sbjct 444 QARDLGKQRDSCLREAEELRTQLR--LLEDARDGLRRELLEAQRKLRESQEG 493
> CE20205
Length=354
Score = 27.3 bits (59), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 32/74 (43%), Gaps = 4/74 (5%)
Query 1 WQDLGIKARELKRQSDRCYRESLQERDKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 60
++DL R L D+ E L + +++ G R F N+ G S++
Sbjct 44 YRDLDAPLRYLSDVPDKIIYEILSKAKLFELFKIQKVSRGFRSFVNNH----HVGFKSIN 99
Query 61 MSSKLEAVASRLDG 74
+S KLE + +DG
Sbjct 100 ISFKLEKIEMLIDG 113
Lambda K H
0.313 0.127 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40