bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2994_orf1
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
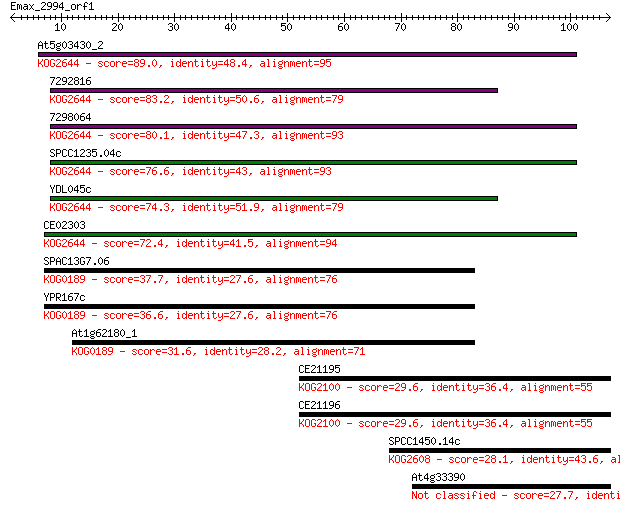
At5g03430_2 89.0 2e-18
7292816 83.2 1e-16
7298064 80.1 9e-16
SPCC1235.04c 76.6 1e-14
YDL045c 74.3 4e-14
CE02303 72.4 2e-13
SPAC13G7.06 37.7 0.006
YPR167c 36.6 0.011
At1g62180_1 31.6 0.36
CE21195 29.6 1.4
CE21196 29.6 1.6
SPCC1450.14c 28.1 4.3
At4g33390 27.7 5.3
> At5g03430_2
Length=447
Score = 89.0 bits (219), Expect = 2e-18, Method: Composition-based stats.
Identities = 46/99 (46%), Positives = 63/99 (63%), Gaps = 6/99 (6%)
Query 6 PI-AFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYC 64
PI A LG R DP A G+E + S W PPF+R+ P+L ++Y +W LL ++ YC
Sbjct 83 PIRAIFLGVRIGDPTAVGQEQFSPS-SPGW-PPFMRVNPILDWSYRDVWAFLLTCKVKYC 140
Query 65 SLYDQGYTSIGNMENTRPNPLLFIK---KEKKYLPAYKL 100
SLYDQGYTSIG++ +T PN LL + ++K+ PAY L
Sbjct 141 SLYDQGYTSIGSIHDTVPNSLLSVNDTSSKEKFKPAYLL 179
> 7292816
Length=294
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/79 (50%), Positives = 53/79 (67%), Gaps = 2/79 (2%)
Query 8 AFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSLY 67
A +G+RNTDP Q L P+Q + N PP +R+ PLL+++Y +W + +PYCSLY
Sbjct 174 AVFVGSRNTDPYCQ--HLAPMQPTDNDWPPMMRLNPLLEWSYHDVWHYIHLNSVPYCSLY 231
Query 68 DQGYTSIGNMENTRPNPLL 86
D+GYTSIGN NT PNP L
Sbjct 232 DRGYTSIGNRANTVPNPHL 250
> 7298064
Length=254
Score = 80.1 bits (196), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 44/97 (45%), Positives = 58/97 (59%), Gaps = 6/97 (6%)
Query 8 AFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSLY 67
A LG R +DP + LY ++ ++N P +RI PLL+++Y IW L LPYC LY
Sbjct 137 AIFLGCRRSDP--ESCNLYELEPTNNGWPAMMRIFPLLEWSYHDIWNYLRSNNLPYCCLY 194
Query 68 DQGYTSIGNMENTRPNPLLFIKKEK----KYLPAYKL 100
DQGYTS+G+ +TR NP L EK Y PAY+L
Sbjct 195 DQGYTSLGDRSSTRVNPSLLAYDEKLDKMTYRPAYEL 231
> SPCC1235.04c
Length=265
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 55/93 (59%), Gaps = 3/93 (3%)
Query 8 AFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSLY 67
A ++G R DP G +++ P F+RIQP+L ++Y IW LLL+ YCSLY
Sbjct 138 AILIGIRRLDP--HGLHRIAFEVTDKGWPKFMRIQPILDWSYTEIWDLLLETNTKYCSLY 195
Query 68 DQGYTSIGNMENTRPNPLLFIKKEKKYLPAYKL 100
D+GYTS+G + +T PNP L + Y PAY L
Sbjct 196 DRGYTSLGGVSDTSPNPAL-KNPDGTYSPAYLL 227
> YDL045c
Length=306
Score = 74.3 bits (181), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 52/80 (65%), Gaps = 4/80 (5%)
Query 8 AFVLGTRNTDPCAQGEELYPIQLS-SNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSL 66
A V+G R+TDP GE L PIQ + SNW P F+R+QPLL + +IW LL P C L
Sbjct 160 AIVIGIRHTDPF--GEALKPIQRTDSNW-PDFMRLQPLLHWDLTNIWSFLLYSNEPICGL 216
Query 67 YDQGYTSIGNMENTRPNPLL 86
Y +G+TSIG + N+ PNP L
Sbjct 217 YGKGFTSIGGINNSLPNPHL 236
> CE02303
Length=519
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 58/97 (59%), Gaps = 4/97 (4%)
Query 7 IAFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSL 66
I ++G+R TDP + + P++ + + P LR+ P+L + Y +W +L +PYC L
Sbjct 411 IPVLMGSRATDPNGKYMKT-PVEWTDSDWPQVLRVCPILNWTYTDVWHMLRGLCVPYCKL 469
Query 67 YDQGYTSIGNMENTRPNPLLFI---KKEKKYLPAYKL 100
YDQGYTS+G +NT +P L I + YLPAYKL
Sbjct 470 YDQGYTSLGGRDNTVKHPALRIVSSDGREHYLPAYKL 506
> SPAC13G7.06
Length=266
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 39/76 (51%), Gaps = 5/76 (6%)
Query 7 IAFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSL 66
+A G R + +G L +QL P L+I PL +++ + ++ +PY L
Sbjct 151 LAVFTGRRRSQGGERGS-LPIVQLDG----PVLKINPLANWSFTEVHNYIITNNVPYNEL 205
Query 67 YDQGYTSIGNMENTRP 82
++GY S+G+ +T+P
Sbjct 206 LNKGYRSVGDWHSTQP 221
> YPR167c
Length=261
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 36/76 (47%), Gaps = 2/76 (2%)
Query 7 IAFVLGTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSL 66
I+ V R + +L I++ L L+I PL+ + + + + +PY L
Sbjct 152 ISAVFTGRRKSQGSARSQLSIIEIDE--LNGILKINPLINWTFEQVKQYIDANNVPYNEL 209
Query 67 YDQGYTSIGNMENTRP 82
D GY SIG+ +T+P
Sbjct 210 LDLGYRSIGDYHSTQP 225
> At1g62180_1
Length=320
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 32/71 (45%), Gaps = 2/71 (2%)
Query 12 GTRNTDPCAQGEELYPIQLSSNWLPPFLRIQPLLQFAYGHIWWLLLQQQLPYCSLYDQGY 71
GTR+ P Q + ++ + + ++ PL +W L +P +L+ QGY
Sbjct 224 GTRSEIPIVQVDPVF--EGLDGGVGSLVKWNPLANVEGADVWNFLRTMDVPVNALHAQGY 281
Query 72 TSIGNMENTRP 82
SIG TRP
Sbjct 282 VSIGCEPCTRP 292
> CE21195
Length=779
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 27/62 (43%), Gaps = 8/62 (12%)
Query 52 IWWLLLQQQLPYCSLYDQGYT---SIGNMENTRPNPL----LFIKKEKKYLPAYKLFLWH 104
+WW QL Y S YD T S+ P P+ + K K LP Y L +W+
Sbjct 226 MWWSTKGDQLAYAS-YDNHLTKNVSLKTYHRLEPYPIDTNFHYPKTFAKVLPTYTLSIWN 284
Query 105 KE 106
K+
Sbjct 285 KK 286
> CE21196
Length=799
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 27/62 (43%), Gaps = 8/62 (12%)
Query 52 IWWLLLQQQLPYCSLYDQGYT---SIGNMENTRPNPL----LFIKKEKKYLPAYKLFLWH 104
+WW QL Y S YD T S+ P P+ + K K LP Y L +W+
Sbjct 246 MWWSTKGDQLAYAS-YDNHLTKNVSLKTYHRLEPYPIDTNFHYPKTFAKVLPTYTLSIWN 304
Query 105 KE 106
K+
Sbjct 305 KK 306
> SPCC1450.14c
Length=571
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 68 DQGYTSIGNMENTRPNPLLFIKKEKK-YLPAYKLFLWHKE 106
+ Y+ I N+ N+R +PLLF EK Y+ Y+L L++KE
Sbjct 53 NSSYSDIENL-NSRVSPLLFDLTEKSDYMRFYRLNLFNKE 91
> At4g33390
Length=779
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 72 TSIGNMENTRPNPLLF-IKKEKKYLPAYKLFLWHKE 106
TS+ N T P P + +KK+KK P + +FL K+
Sbjct 741 TSVSNETETNPIPQVNPVKKKKKLFPRFFMFLMKKK 776
Lambda K H
0.322 0.142 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167556980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40