bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2966_orf2
Length=211
Score E
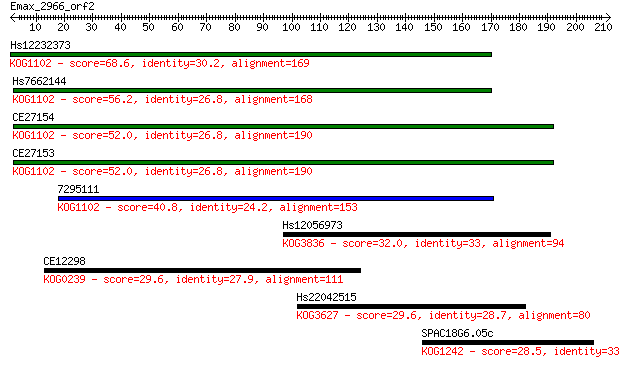
Sequences producing significant alignments: (Bits) Value
Hs12232373 68.6 1e-11
Hs7662144 56.2 4e-08
CE27154 52.0 9e-07
CE27153 52.0 1e-06
7295111 40.8 0.002
Hs12056973 32.0 0.85
CE12298 29.6 4.9
Hs22042515 29.6 5.0
SPAC18G6.05c 28.5 9.9
> Hs12232373
Length=997
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 51/169 (30%), Positives = 83/169 (49%), Gaps = 23/169 (13%)
Query 1 CRKEIEKSECELAQKRKIIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTF 60
CR+E++K+E E+ + II +YKQIC + E ++ V++ +IR K+ DCE C F
Sbjct 844 CRRELDKAESEIKKNSSIIGDYKQICSQLSERLEKQQTANKVEIEKIRQKVDDCERCREF 903
Query 61 LTELNNNGRDSNGETTSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKVALAE 120
N GR +T DT ++ K+ L ++ ++E +L +TK+ L E
Sbjct 904 ---FNKEGRVKGISSTKEVLDEDT---------DEEKETLKNQLREMELELAQTKLQLVE 951
Query 121 AEDRNGFLAGKLHTTSSELSLIKAQVEEAAHQKQNTWITKALSSIRDST 169
AE K+ L L +V+ A + TW + LSSI+ +T
Sbjct 952 AE-------CKIQDLEHHLGLALNEVQAA----KKTWFNRTLSSIKTAT 989
> Hs7662144
Length=370
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 45/183 (24%), Positives = 83/183 (45%), Gaps = 53/183 (28%)
Query 2 RKEIEKSECELAQKRKIIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTFL 61
RK++EK+E E+ + II EYKQIC + E ++ + ++ ++ K+ C+ CS
Sbjct 208 RKQLEKAEYEIKKTTAIIAEYKQICSQLSTRLEKQQAASKEELEVVKGKMMACKHCSDIF 267
Query 62 T-----ELNNNGRDSNGETTSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKV 116
+ +L GR+ G E++ KD L +++ ++E +L +TK+
Sbjct 268 SKEGALKLAATGREDQG-----------------IETDDEKDSLKKQLREMELELAQTKL 310
Query 117 ALAEAEDRNGFLAGKLHTTSSELSLIKAQVEEAAHQK----------QNTWITKALSSIR 166
L EA K +++E HQ+ +N+W +K L+SI+
Sbjct 311 QLVEA---------------------KCKIQELEHQRGALMNEIQAAKNSWFSKTLNSIK 349
Query 167 DST 169
+T
Sbjct 350 TAT 352
> CE27154
Length=1140
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 51/191 (26%), Positives = 92/191 (48%), Gaps = 28/191 (14%)
Query 2 RKEIEKSECELAQKRKIIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTFL 61
R+++ + E ++ K++ EYK++ + E+ERE V I +I+DC+ C +
Sbjct 815 RRDVLRLEENGSRAEKLLAEYKKLFSERSKRAENEREHFEVQKKAIIARISDCDKCWPAV 874
Query 62 TELNNNGRDSNGETTSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKVALAEA 121
E N + +T +GP ++T+ + +D I +LE DL +TK++L EA
Sbjct 875 CEWEKNRSPVHSASTPTGP-------DLLTKLEERED----HIKNLEIDLAQTKLSLVEA 923
Query 122 EDRNGFLAGKLHTTSSELSLIKAQVEEAAHQKQNTWITKALSSIRDSTTGSAVSKV-KSS 180
E RN L +L AQ E + W K ++ +++ GS++ +S+
Sbjct 924 ECRNQDLTHQL----------MAQSESDGKK----WFKKTITQLKE--VGSSLKHHERSN 967
Query 181 SSVSSGISSSM 191
SSV+ SS+
Sbjct 968 SSVTPHFSSTF 978
> CE27153
Length=1136
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 51/191 (26%), Positives = 92/191 (48%), Gaps = 28/191 (14%)
Query 2 RKEIEKSECELAQKRKIIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTFL 61
R+++ + E ++ K++ EYK++ + E+ERE V I +I+DC+ C +
Sbjct 811 RRDVLRLEENGSRAEKLLAEYKKLFSERSKRAENEREHFEVQKKAIIARISDCDKCWPAV 870
Query 62 TELNNNGRDSNGETTSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKVALAEA 121
E N + +T +GP ++T+ + +D I +LE DL +TK++L EA
Sbjct 871 CEWEKNRSPVHSASTPTGP-------DLLTKLEERED----HIKNLEIDLAQTKLSLVEA 919
Query 122 EDRNGFLAGKLHTTSSELSLIKAQVEEAAHQKQNTWITKALSSIRDSTTGSAVSKV-KSS 180
E RN L +L AQ E + W K ++ +++ GS++ +S+
Sbjct 920 ECRNQDLTHQL----------MAQSESDGKK----WFKKTITQLKE--VGSSLKHHERSN 963
Query 181 SSVSSGISSSM 191
SSV+ SS+
Sbjct 964 SSVTPHFSSTF 974
> 7295111
Length=1194
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/156 (23%), Positives = 72/156 (46%), Gaps = 19/156 (12%)
Query 18 IIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTFLTELNNNGRDSNGET-- 75
II+EYKQI + + + E + ++ +++C+ C + N+N + +G+
Sbjct 962 IIQEYKQIIQRQEQDMNTLSET----LGKVMHMVSNCQDCQQQIDAGNDNAKSDDGKNRG 1017
Query 76 -TSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKVALAEAEDRNGFLAGKLHT 134
T + ++ P+ + N +RI +LE +L + K+A EAE +N L +L
Sbjct 1018 QTDAMRNANEHPLGPLDPLNAAS----QRIRELELELAQAKLAQVEAECKNQDLNHQLSN 1073
Query 135 TSSELSLIKAQVEEAAHQKQNTWITKALSSIRDSTT 170
T SEL + W++K +S+++ T
Sbjct 1074 TLSEL--------QTNRNSWQPWLSKTFNSLQEKVT 1101
> Hs12056973
Length=306
Score = 32.0 bits (71), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 13/103 (12%)
Query 97 KDLLLRRIADLESDLIKTK-----VALAEAEDRNGFLAGKLHTTSSELSLIKAQVEEAAH 151
K++LL+R ADL L T + L A D +A L++ S + A AH
Sbjct 97 KEMLLKRAADLVEALYGTPHNNQDIILKRAAD----IAEALYSVPRNPSQLPALSSSPAH 152
Query 152 QKQ---NTWITKALSSIRDSTTGSAVSKVKSSSSVSS-GISSS 190
N++ ++ SI +ST G+ ++++SS+S G SSS
Sbjct 153 SGMMGINSYGSQLGVSISESTQGNNQGYIRNTSSISPRGYSSS 195
> CE12298
Length=587
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 43/111 (38%), Gaps = 8/111 (7%)
Query 13 AQKRKIIEEYKQICKNYQCKFESERERAIVDMAQIRDKITDCEACSTFLTELNNNGRDSN 72
A R + E + K C R I M +I+ A STF +L GR
Sbjct 68 ATGRASLPERSAMAKPASC------SRPIPTMQSTASRISTLTAASTF-RQLRT-GRPPP 119
Query 73 GETTSSGPPSDTKPITVVTESNKYKDLLLRRIADLESDLIKTKVALAEAED 123
T S S KP K +L ++A LE + + + A+AEA D
Sbjct 120 PSTQRSTATSSLKPSVTRARPVAQKPILPSKMALLEEKIAQLESAMAEATD 170
> Hs22042515
Length=280
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 1/81 (1%)
Query 102 RRIADLESDLIKTKVA-LAEAEDRNGFLAGKLHTTSSELSLIKAQVEEAAHQKQNTWITK 160
R +A LE + +V L AE L +L T LSL+ Q+ E AH Q +
Sbjct 177 RLLAALEEEACPARVGPLQPAESEGSALVLRLPQTLGALSLLLGQLSEVAHALQGHVVAV 236
Query 161 ALSSIRDSTTGSAVSKVKSSS 181
+ R+ GS V ++
Sbjct 237 EMEDEREVGKGSPQLPVDQAA 257
> SPAC18G6.05c
Length=2670
Score = 28.5 bits (62), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 33/60 (55%), Gaps = 3/60 (5%)
Query 146 VEEAAHQKQNTWITKALSSIRDSTTGSAVSKVKSSSSVSSGISSSMVDGHQNSNSHVFNN 205
+E+A + + LSS+ G A SKV +SS + S + +S V+G ++SN+ V N
Sbjct 282 IEKALLRSPEIIFSGILSSL---AHGFADSKVDASSLILSSVLTSFVNGLKSSNAEVRRN 338
Lambda K H
0.307 0.121 0.326
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3825978242
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40