bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2931_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
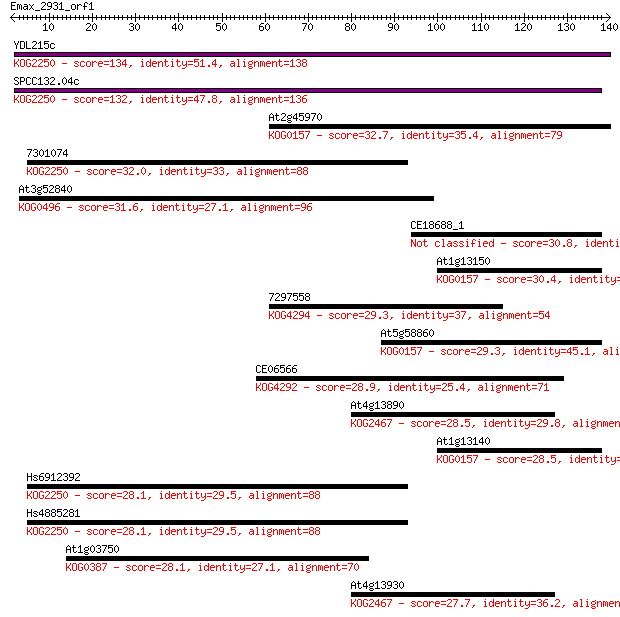
YDL215c 134 4e-32
SPCC132.04c 132 2e-31
At2g45970 32.7 0.19
7301074 32.0 0.35
At3g52840 31.6 0.48
CE18688_1 30.8 0.88
At1g13150 30.4 1.0
7297558 29.3 2.5
At5g58860 29.3 2.7
CE06566 28.9 3.2
At4g13890 28.5 3.9
At1g13140 28.5 4.1
Hs6912392 28.1 5.0
Hs4885281 28.1 5.2
At1g03750 28.1 5.3
At4g13930 27.7 6.3
> YDL215c
Length=1092
Score = 134 bits (338), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 71/140 (50%), Positives = 92/140 (65%), Gaps = 7/140 (5%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
WWK+F TGK GGIPHD +GMT+ V A+V IY LNL + + QTGGPDGDLG
Sbjct 701 WWKSFLTGKSPSLGGIPHDEYGMTSLGVRAYVNKIYETLNLTNSTVYKFQTGGPDGDLGS 760
Query 62 NALLQTKSKT--IAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQG 119
N +L + +A++D SGVL DPKGL+K+E LCRL E K + +++S LS G
Sbjct 761 NEILLSSPNECYLAILDGSGVLCDPKGLDKDE---LCRLAHERKMISD--FDTSKLSNNG 815
Query 120 FKVAQDARDIILPDGTFVAS 139
F V+ DA DI+LP+GT VA+
Sbjct 816 FFVSVDAMDIMLPNGTIVAN 835
> SPCC132.04c
Length=1106
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 65/137 (47%), Positives = 91/137 (66%), Gaps = 6/137 (4%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKE-EEMTRIQTGGPDGDLG 60
WWK+F TGK GGIPHD++GMT+ SV +V+GIY KLN+ + ++T++QTGGPDGDLG
Sbjct 726 WWKSFFTGKKPTMGGIPHDKYGMTSLSVRCYVEGIYKKLNITDPSKLTKVQTGGPDGDLG 785
Query 61 CNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGF 120
N + + K IAVID SGVLYDP GL++ EL LR + +++ LSP+G+
Sbjct 786 SNEIKLSNEKYIAVIDGSGVLYDPAGLDRTEL-----LRLADERKTIDHFDAGKLSPEGY 840
Query 121 KVAQDARDIILPDGTFV 137
+V ++ LP+G V
Sbjct 841 RVLVKDTNLKLPNGEIV 857
> At2g45970
Length=537
Score = 32.7 bits (73), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 13/79 (16%)
Query 61 CNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGF 120
C L++T+ +A L+ + L+ EEL RL L+ ET LY S P+
Sbjct 336 CTVLVETRGDDVA-------LWTDEPLSCEELDRLVFLKAALSET-LRLYPSV---PEDS 384
Query 121 KVAQDARDIILPDGTFVAS 139
K A +D +LPDGTFV +
Sbjct 385 KRA--VKDDVLPDGTFVPA 401
> 7301074
Length=590
Score = 32.0 bits (71), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 44/97 (45%), Gaps = 10/97 (10%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPD------GD 58
A TGK I GGI H R T V ++ N+ N + T GG G+
Sbjct 246 ACVTGKPINQGGI-HGRVSATGRGVFHGLENFINEANYMSQIGTTPGWGGKTFIVQGFGN 304
Query 59 LGCNA---LLQTKSKTIAVIDASGVLYDPKGLNKEEL 92
+G + L + + I VI+ G LY+P+G++ + L
Sbjct 305 VGLHTTRYLTRAGATCIGVIEHDGTLYNPEGIDPKLL 341
> At3g52840
Length=727
Score = 31.6 bits (70), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 42/97 (43%), Gaps = 7/97 (7%)
Query 3 WKAFTTGKLIQHGG-IPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
W TG + GG IP+ SV F++ + +N + GG + D
Sbjct 252 WTENWTGWFTEFGGAIPNRPVEDIAFSVARFIQNGGSFMNY------YMYYGGTNFDRTA 305
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRL 98
+ T A ID G+L +PK + +ELH++ +L
Sbjct 306 GVFIATSYDYDAPIDEYGLLREPKYSHLKELHKVIKL 342
> CE18688_1
Length=679
Score = 30.8 bits (68), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 2/44 (4%)
Query 94 RLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDARDIILPDGTFV 137
R+C++ EG NA+ Y SSL P+ D+ L TFV
Sbjct 532 RMCKIDHEG--YNALTYQSSLRPPRAIDFIDFPNDVTLDGPTFV 573
> At1g13150
Length=529
Score = 30.4 bits (67), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 7/45 (15%)
Query 100 FEGKETNAMLYNSSLLS-------PQGFKVAQDARDIILPDGTFV 137
F KE N M+Y + LS P ++ Q D +LPDGTFV
Sbjct 365 FTVKELNNMVYLQAALSETLRLFPPIPMEMKQAIEDDVLPDGTFV 409
> 7297558
Length=1043
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 5/54 (9%)
Query 61 CNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSL 114
CN +KSK ++++D V P G+N+ ELH L L +T M+ N SL
Sbjct 645 CNFNKISKSKALSLLDQMQV---PVGINRSELHNLFNLTLLPVDT--MISNDSL 693
> At5g58860
Length=513
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Query 87 LNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDARDIILPDGTFV 137
L +E RL L+ ET LY S PQ FK D D +LPDGTFV
Sbjct 353 LEFDEADRLVYLKAALAET-LRLYPSV---PQDFKYVVD--DDVLPDGTFV 397
> CE06566
Length=3871
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 30/71 (42%), Gaps = 5/71 (7%)
Query 58 DLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSP 117
D C L+ ++ ++ D K +NKE H+ CR +G +T ++ N
Sbjct 3510 DSDCGGWLKATNEIKTLVYKGITSDDNKEMNKERSHQRCRFMIQGPKTEPVIVNF----- 3564
Query 118 QGFKVAQDARD 128
Q F + A D
Sbjct 3565 QQFNIPSKAGD 3575
> At4g13890
Length=470
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 80 VLYD--PKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDA 126
+L+D P GL ++ ++C L + NA+ ++S L+P G ++ A
Sbjct 346 ILWDLRPLGLTGNKVEKVCELCYITLNRNAVFGDTSFLAPGGVRIGTPA 394
> At1g13140
Length=519
Score = 28.5 bits (62), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query 100 FEGKETNAMLYNSSLLS-------PQGFKVAQDARDIILPDGTFV 137
F KE N M+Y + LS P ++ Q D + PDGTF+
Sbjct 357 FTVKELNDMVYLQAALSETMRLYPPIPMEMKQAIEDDVFPDGTFI 401
> Hs6912392
Length=558
Score = 28.1 bits (61), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 10/97 (10%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLK---------EEEMTRIQTGGP 55
A TGK I GGI H R T V ++ N+ + ++ +Q G
Sbjct 253 ACVTGKPISQGGI-HGRISATGRGVFHGIENFINQASYMSILGMTPGFRDKTFVVQGFGN 311
Query 56 DGDLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEEL 92
G L + +K IAV ++ G +++P G++ +EL
Sbjct 312 VGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> Hs4885281
Length=558
Score = 28.1 bits (61), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 10/97 (10%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKE---------EEMTRIQTGGP 55
A TGK I GGI H R T V ++ N+ + ++ +Q G
Sbjct 253 ACVTGKPISQGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTFVVQGFGN 311
Query 56 DGDLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEEL 92
G L + +K IAV ++ G +++P G++ +EL
Sbjct 312 VGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> At1g03750
Length=874
Score = 28.1 bits (61), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 31/71 (43%), Gaps = 17/71 (23%)
Query 14 HGGIPHDRFGM-TTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGCNALLQTKSKTI 72
HGGI D G+ T AF+ +Y G DGD G + LL++ +
Sbjct 170 HGGILGDDMGLGKTIQTIAFLAAVY----------------GKDGDAGESCLLESDKGPV 213
Query 73 AVIDASGVLYD 83
+I S ++++
Sbjct 214 LIICPSSIIHN 224
> At4g13930
Length=471
Score = 27.7 bits (60), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 80 VLYD--PKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDA 126
VL+D P GL ++ +LC L NA+ +SS L+P G ++ A
Sbjct 346 VLWDLRPLGLTGNKVEKLCDLCSITLNKNAVFGDSSALAPGGVRIGAPA 394
Lambda K H
0.318 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40