bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2910_orf1
Length=176
Score E
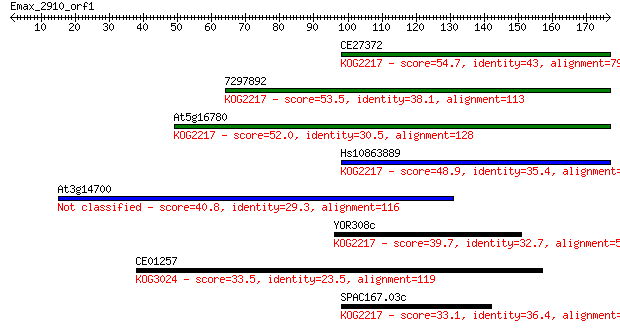
Sequences producing significant alignments: (Bits) Value
CE27372 54.7 9e-08
7297892 53.5 2e-07
At5g16780 52.0 6e-07
Hs10863889 48.9 5e-06
At3g14700 40.8 0.001
YOR308c 39.7 0.003
CE01257 33.5 0.21
SPAC167.03c 33.1 0.28
> CE27372
Length=829
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 50/81 (61%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKI-KKLELERRLQENIMDC-L 155
+VNI Y D G M AKDA+R +S+ FHG+ P ++ EK+ +K + ER L+ N D L
Sbjct 738 DVNISYVDRKGREMDAKDAYRELSYKFHGRNPGKKQLEKRANRKDKEERMLKTNSYDTPL 797
Query 156 PTLKALKKVQEGESSAHLVLT 176
TL +K Q+ S+ +LVL+
Sbjct 798 GTLDKQRKKQKQLSTPYLVLS 818
> 7297892
Length=964
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 68/132 (51%), Gaps = 26/132 (19%)
Query 64 LQYLQSKDFFSLDK-------IRRRRGHHLEQPLHNADNDKE-----VNIDYRDAFGNVM 111
+ +LQ+K++ DK + RR H P+ + DKE V +DY D G ++
Sbjct 822 MAHLQAKNYSIEDKAAGEDEKVGRRDRFHF-GPITDF-KDKETFKPNVKLDYIDDNGRIL 879
Query 112 TAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDC-------LPTLKALKKV 164
K+AFR +S FHGK P K EK++KK+E Q+ +M L TL L++
Sbjct 880 NLKEAFRYLSHKFHGKGPGKNKIEKRLKKME-----QDGLMKTMSSTDTPLGTLTMLQQK 934
Query 165 QEGESSAHLVLT 176
Q+ +A++VL+
Sbjct 935 QKETKTAYVVLS 946
> At5g16780
Length=820
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 39/146 (26%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query 49 EFMAEQPIGDSVSGALQYLQSKDFF---------SLDKIRRR-------RGHHLEQPLHN 92
E + E +G +SGAL+ L+ + ++DK + + G + +
Sbjct 616 ENIHEVAVGKGLSGALKLLKDRGTLKEKVEWGGRNMDKKKSKLVGIVDDDGGKESKDKES 675
Query 93 ADNDKEVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIM 152
D K++ I+ D FG +T K+AFR +S FHGK P K+EK++K+ + E +L++
Sbjct 676 KDRFKDIRIERTDEFGRTLTPKEAFRLLSHKFHGKGPGKMKEEKRMKQYQEELKLKQMKN 735
Query 153 DCLP--TLKALKKVQEGESSAHLVLT 176
P +++ +++ Q + +LVL+
Sbjct 736 SDTPSQSVQRMREAQAQLKTPYLVLS 761
> Hs10863889
Length=800
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 49/81 (60%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLP- 156
+V I+Y D G +T K+AFR++S FHGK K E+++KKL+ E L++ P
Sbjct 707 DVKIEYVDETGRKLTPKEAFRQLSHRFHGKGSGKMKTERRMKKLDEEALLKKMSSSDTPL 766
Query 157 -TLKALKKVQEGESSAHLVLT 176
T+ L++ Q+ + + ++VL+
Sbjct 767 GTVALLQEKQKAQKTPYIVLS 787
> At3g14700
Length=204
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 57/122 (46%), Gaps = 17/122 (13%)
Query 15 SSKKQREETEEGEVVENVPKEDEEEEEEEGDSEPEFMAEQPIGDSVSGALQYLQSKDFFS 74
SS+++RE + E + V K + GD M E +G +SGAL L+ + F
Sbjct 52 SSERRREVCSKAEDI--VDKAIDNHSRVRGDG---IMREADVGTGLSGALNRLREQGTF- 105
Query 75 LDKIRRRRGHHLEQPLHNADND------KEVNIDYRDAFGNVMTAKDAFRRISWHFHGKF 128
+ G + +N ++D K++ I + +G +MT K+A+R + FHGK
Sbjct 106 -----KEEGKVVGVKDNNHEDDRFKDRFKDIQIQRVNKWGRIMTEKEAYRSLCHGFHGKG 160
Query 129 PS 130
P
Sbjct 161 PG 162
> YOR308c
Length=587
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 96 DKEVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQEN 150
D ++ + YRD GN +T K+A++++S FHG + +K+ K ++E + EN
Sbjct 524 DPDIKLVYRDEKGNRLTTKEAYKKLSQKFHGTKSNKKKRAKMKSRIEARKNTPEN 578
> CE01257
Length=362
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 51/119 (42%), Gaps = 6/119 (5%)
Query 38 EEEEEEGDSEPEFMAEQPIGDSVSGALQYLQSKDFFSLDKIRRRRGHHLEQPLHNADNDK 97
E E E G+SEPE + V A+Q+ + K + I ++ G H K
Sbjct 100 EIENEAGNSEPEQLLNTARCKCVDLAIQWTKQK---ATTPIEKKYG---SAAFHTVLAKK 153
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLP 156
V +D+ + N D F+ + H ++ S +++E + + + +E Q +D P
Sbjct 154 LVAVDHVELAKNHFLLSDDFKSFANFLHQEYESFKQKESEPEVIIVEAVFQSLCLDRFP 212
> SPAC167.03c
Length=649
Score = 33.1 bits (74), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 28/45 (62%), Gaps = 1/45 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWH-FHGKFPSLRKQEKKIKKL 141
+V+I Y D FG + K+A++ + H FHGK K EK+++++
Sbjct 593 QVDIKYVDEFGVELGPKEAYKYLLSHQFHGKGSGKAKTEKRLRRI 637
Lambda K H
0.308 0.127 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40