bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2875_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
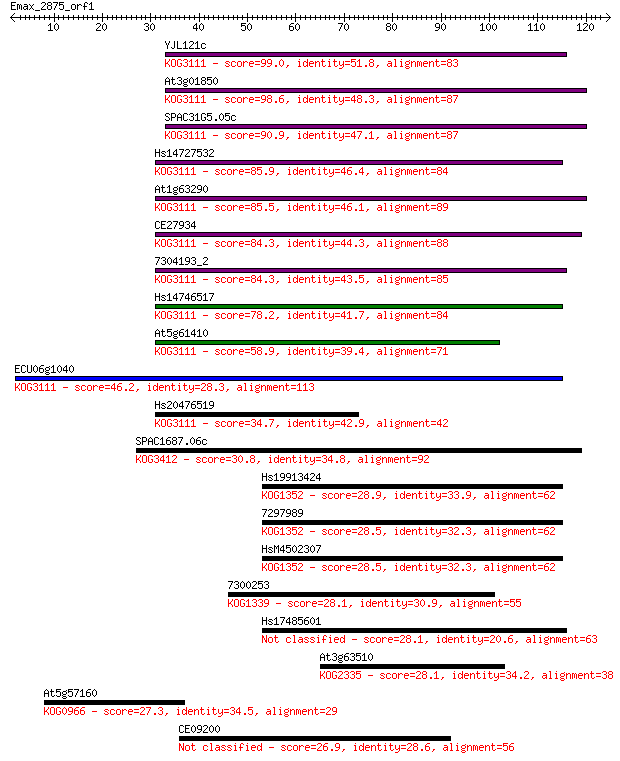
YJL121c 99.0 2e-21
At3g01850 98.6 3e-21
SPAC31G5.05c 90.9 6e-19
Hs14727532 85.9 2e-17
At1g63290 85.5 2e-17
CE27934 84.3 4e-17
7304193_2 84.3 5e-17
Hs14746517 78.2 4e-15
At5g61410 58.9 2e-09
ECU06g1040 46.2 1e-05
Hs20476519 34.7 0.044
SPAC1687.06c 30.8 0.66
Hs19913424 28.9 2.2
7297989 28.5 2.9
HsM4502307 28.5 3.3
7300253 28.1 4.0
Hs17485601 28.1 4.1
At3g63510 28.1 4.2
At5g57160 27.3 7.8
CE09200 26.9 8.2
> YJL121c
Length=238
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 43/83 (51%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 33 IDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGANVI 92
+D+ L+MTVEP FGGQ F+ + M KV+ R FP+LNIQVDGGL ET+ AA++GANVI
Sbjct 145 LDMALVMTVEPGFGGQKFMEDMMPKVETLRAKFPHLNIQVDGGLGKETIPKAAKAGANVI 204
Query 93 VAGTSLYNCPSPGTLMEYMRDVI 115
VAGTS++ P ++ +M++ +
Sbjct 205 VAGTSVFTAADPHDVISFMKEEV 227
> At3g01850
Length=233
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 42/87 (48%), Positives = 62/87 (71%), Gaps = 0/87 (0%)
Query 33 IDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGANVI 92
++++L+MTVEP FGGQ F+ + M+KV+ RQ +P L+IQVDGGL T+ AA +GAN I
Sbjct 144 VEMVLVMTVEPGFGGQKFMPDMMDKVRALRQKYPTLDIQVDGGLGPSTIDTAAAAGANCI 203
Query 93 VAGTSLYNCPSPGTLMEYMRDVIVAAQ 119
VAG+S++ P PG ++ +R + AQ
Sbjct 204 VAGSSVFGAPEPGDVISLLRTSVEKAQ 230
> SPAC31G5.05c
Length=228
Score = 90.9 bits (224), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 41/87 (47%), Positives = 67/87 (77%), Gaps = 0/87 (0%)
Query 33 IDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGANVI 92
+D++L+MTVEP GGQ+F+ E + KV+ R+ +P LN++VDGGL+ +TV AA++GANVI
Sbjct 135 LDMVLVMTVEPGKGGQSFMPECLPKVEFLRKKYPTLNVEVDGGLSLKTVDAAADAGANVI 194
Query 93 VAGTSLYNCPSPGTLMEYMRDVIVAAQ 119
VAGT++++ SP ++ +R+ ++ AQ
Sbjct 195 VAGTAVFHAQSPEEVISGLRNSVMKAQ 221
> Hs14727532
Length=203
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 58/84 (69%), Gaps = 0/84 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGAN 90
N ID+ L+MTVEP FGGQ F+ + M KV R FP L+I+VDGG+ +TV AE+GAN
Sbjct 108 NQIDMALVMTVEPGFGGQKFMEDMMPKVHWLRTQFPSLDIEVDGGVGPDTVHKCAEAGAN 167
Query 91 VIVAGTSLYNCPSPGTLMEYMRDV 114
+IV+G+++ P +++ +R+V
Sbjct 168 MIVSGSAIMRSEDPRSVINLLRNV 191
> At1g63290
Length=227
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/89 (46%), Positives = 61/89 (68%), Gaps = 0/89 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGAN 90
N ++++L+MTVEP FGGQ F+ M+KV+ R +P L+I+VDGGL T+ AA +GAN
Sbjct 136 NPVEMVLVMTVEPGFGGQKFMPSMMDKVRALRNKYPTLDIEVDGGLGPSTIDAAAAAGAN 195
Query 91 VIVAGTSLYNCPSPGTLMEYMRDVIVAAQ 119
IVAG+S++ P PG ++ +R + AQ
Sbjct 196 CIVAGSSVFGAPKPGDVISLLRASVEKAQ 224
> CE27934
Length=227
Score = 84.3 bits (207), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 39/88 (44%), Positives = 55/88 (62%), Gaps = 0/88 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGAN 90
N +D LIMTVEP FGGQ F+ M KV+ R +P L IQVDGG+ E ++++A++GAN
Sbjct 136 NHLDNALIMTVEPGFGGQKFMENMMEKVRTIRSKYPNLTIQVDGGVTPENIEISAQAGAN 195
Query 91 VIVAGTSLYNCPSPGTLMEYMRDVIVAA 118
IV+GT + M +R+ + AA
Sbjct 196 AIVSGTGIIKAADQSVAMTTIRNAVEAA 223
> 7304193_2
Length=221
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 62/85 (72%), Gaps = 0/85 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGAN 90
++ D++L+MTVEP FGGQ+F+ + M KVK R+ +P L+I+VDGG+ +T+ AE+GAN
Sbjct 132 SIADVVLVMTVEPGFGGQSFMADMMPKVKWLRENYPNLDIEVDGGVGPKTIHCCAEAGAN 191
Query 91 VIVAGTSLYNCPSPGTLMEYMRDVI 115
+IV+GT++ +++ +RDV+
Sbjct 192 MIVSGTAVVGASDQSQVIKELRDVV 216
> Hs14746517
Length=228
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGAN 90
N ID+ L+MTVEP FG Q F+ + M KV R FP L+I+ DGG+ ++TV AE+GAN
Sbjct 133 NQIDMALVMTVEPGFGEQKFMEDMMPKVHWLRTQFPSLDIEGDGGVGSDTVHKCAEAGAN 192
Query 91 VIVAGTSLYNCPSPGTLMEYMRDV 114
+ V+G+++ P +++ +R++
Sbjct 193 MTVSGSAIMRSEDPRSVINLLRNI 216
> At5g61410
Length=281
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 48/77 (62%), Gaps = 8/77 (10%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQL------FPYLNIQVDGGLNAETVQVA 84
+++DL+LIM+V P FGGQ+F+ Q+ K+ R++ P+ I+VDGG+
Sbjct 186 DMVDLVLIMSVNPGFGGQSFIESQVKKISDLRKMCAEKGVNPW--IEVDGGVTPANAYKV 243
Query 85 AESGANVIVAGTSLYNC 101
E+GAN +VAG++++
Sbjct 244 IEAGANALVAGSAVFGA 260
> ECU06g1040
Length=217
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/116 (27%), Positives = 55/116 (47%), Gaps = 10/116 (8%)
Query 2 CTWLNCYSYLKSCCFYFFTYINSSSSSSNNLI---DLLLIMTVEPEFGGQTFLYEQMNKV 58
C YL+ F +N + + + D +LIM+V+P FGGQ F E + KV
Sbjct 98 CDKAGVAEYLRKRNVLFGIALNPETQVDDAEMRSADFVLIMSVKPGFGGQKFQEECLAKV 157
Query 59 KKARQLFPYLNIQVDGGLNAETVQVAAESGANVIVAGTSLYNCPSPGTLMEYMRDV 114
++ R+ + I DGG+ E + +GA+ V G+ + G +++RD+
Sbjct 158 EEVRRYGKMVGI--DGGI--EMSNIGRITGADYAVVGSGYFR---SGDRKKFLRDI 206
> Hs20476519
Length=118
Score = 34.7 bits (78), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 31 NLIDLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQV 72
N ID+ L+MTVEP F Q F+ + M KV + L+I+V
Sbjct 74 NKIDMALVMTVEPGFEEQKFMKDMMPKVHWLQTQLLSLDIEV 115
> SPAC1687.06c
Length=134
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 41/98 (41%), Gaps = 15/98 (15%)
Query 27 SSSNNLI-----DLLLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETV 81
S SN+LI D + PEFGG F E +N K Q F L N + V
Sbjct 2 SVSNDLIWQVIRDNNRFLVKRPEFGGIQFNREPVNVSGKNAQRFSGL-------CNDKAV 54
Query 82 QVAAESGANVI-VAGTSLYNCPSPGTLMEYMRDVIVAA 118
V A S V+ + T+ N P L + +DVI A
Sbjct 55 GVQANSPRGVVLITKTNPKNAQKPAKL--FRKDVIANA 90
> Hs19913424
Length=617
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 53 EQMNKVKKARQLFPYLNIQVDGGLNA---ETVQVAAESGANVIVAGTSLYNCPSPGTLME 109
E+ N++ + + FP L ++VDG + + T VA S V S+Y + TL E
Sbjct 279 ERGNEMSEVLRDFPELTMEVDGKVESIMKRTALVANTSNMPVAAREASIY---TGITLSE 335
Query 110 YMRDV 114
Y RD+
Sbjct 336 YFRDM 340
> 7297989
Length=614
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 53 EQMNKVKKARQLFPYLNIQVDG---GLNAETVQVAAESGANVIVAGTSLYNCPSPGTLME 109
E+ N++ + + FP L++++DG + T VA S V S+Y + TL E
Sbjct 276 ERGNEMSEVLRDFPELSVEIDGVTESIMKRTALVANTSNMPVAAREASIY---TGITLSE 332
Query 110 YMRDV 114
Y RD+
Sbjct 333 YFRDM 337
> HsM4502307
Length=615
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 53 EQMNKVKKARQLFPYLNIQVDG---GLNAETVQVAAESGANVIVAGTSLYNCPSPGTLME 109
E+ N++ + + FP L++++DG + T VA S V S+Y + TL E
Sbjct 277 ERGNEMSEVLRDFPQLSLEIDGVTESIMKRTALVANTSNMPVAAREASIY---TGITLSE 333
Query 110 YMRDV 114
Y RD+
Sbjct 334 YFRDM 338
> 7300253
Length=465
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query 46 GGQTFLYEQMNKVKKARQLFPYLNIQVDGGLNAETVQVAAESGANVIVAGTSLYN 100
G ++ Y + K K F +I V G + +VQ +SG ++I A T++YN
Sbjct 315 GSNSYTYTPVTK--KGYWQFTLQDIYVGGTKVSGSVQAIVDSGTSLITAPTAIYN 367
> Hs17485601
Length=612
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 13/66 (19%), Positives = 34/66 (51%), Gaps = 3/66 (4%)
Query 53 EQMNKVKKARQLFPYLNIQV---DGGLNAETVQVAAESGANVIVAGTSLYNCPSPGTLME 109
+++N V+K + + + QV G +++ ++ + G N++V C PG+++
Sbjct 442 QRVNAVRKLQAAYAQVEAQVYKGPGTISSTGCEICWKEGKNIMVKNIKKQKCEGPGSVVL 501
Query 110 YMRDVI 115
R+++
Sbjct 502 TSRNIL 507
> At3g63510
Length=456
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 65 FPYLNIQVDGGLNA-ETVQVAAESGANVIVAGTSLYNCP 102
FP L ++GG+ + V A + GA+ ++ G + YN P
Sbjct 285 FPDLRFTINGGITSVSKVNAALKEGAHGVMVGRAAYNNP 323
> At5g57160
Length=1184
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 8 YSYLKSCCFYFFTYINSSSSSSNNLIDLL 36
+S L SCC YF+ Y + S+ L+ ++
Sbjct 766 WSCLLSCCVYFYPYSQTLSTEEEALLGIM 794
> CE09200
Length=331
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 27/59 (45%), Gaps = 3/59 (5%)
Query 36 LLIMTVEPEFGGQTFLYEQMNKVKKARQLFPYLNIQV---DGGLNAETVQVAAESGANV 91
L+I + E+ + + E +NK + A + FP I + DG L V +G +V
Sbjct 150 LMIFGLSDEYSDEYMMVEMLNKYELAIKDFPRFIIVIYGADGSLRYRNVMYLVSAGVDV 208
Lambda K H
0.320 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40