bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2873_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
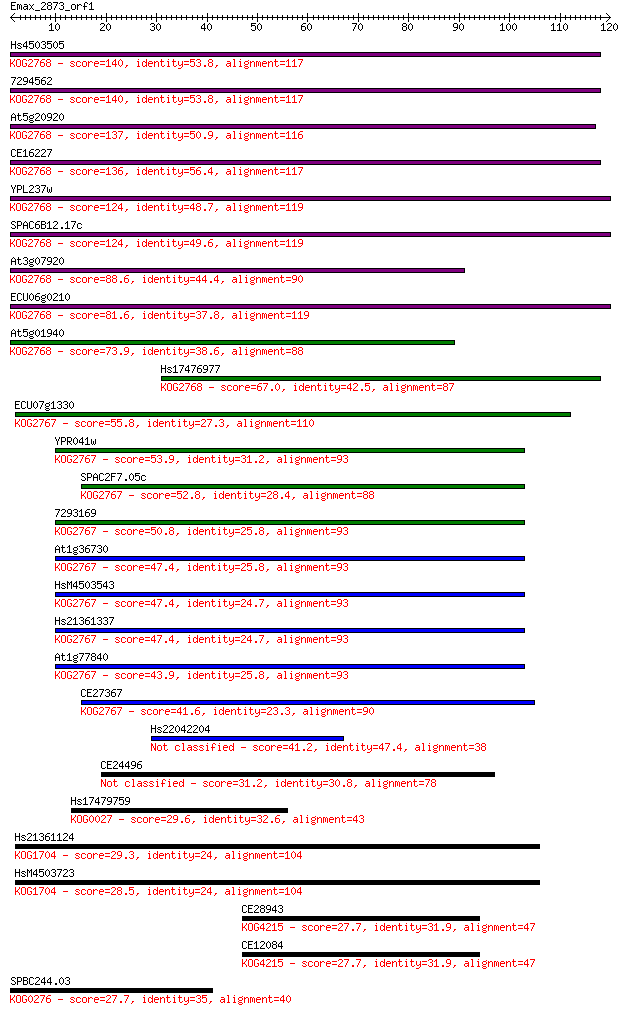
Hs4503505 140 5e-34
7294562 140 5e-34
At5g20920 137 6e-33
CE16227 136 1e-32
YPL237w 124 3e-29
SPAC6B12.17c 124 6e-29
At3g07920 88.6 3e-18
ECU06g0210 81.6 3e-16
At5g01940 73.9 6e-14
Hs17476977 67.0 7e-12
ECU07g1330 55.8 2e-08
YPR041w 53.9 7e-08
SPAC2F7.05c 52.8 2e-07
7293169 50.8 6e-07
At1g36730 47.4 6e-06
HsM4503543 47.4 7e-06
Hs21361337 47.4 7e-06
At1g77840 43.9 7e-05
CE27367 41.6 4e-04
Hs22042204 41.2 5e-04
CE24496 31.2 0.43
Hs17479759 29.6 1.3
Hs21361124 29.3 2.1
HsM4503723 28.5 2.8
CE28943 27.7 4.9
CE12084 27.7 4.9
SPBC244.03 27.7 5.5
> Hs4503505
Length=333
Score = 140 bits (354), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 63/117 (53%), Positives = 86/117 (73%), Gaps = 2/117 (1%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
PQVVRVG+KK +++NF DIC ++HR +H+L F+LAELGT GSI G+ QLV+KG++ K
Sbjct 207 PQVVRVGTKKTSFVNFTDICKLLHRQPKHLLAFLLAELGTSGSIDGNNQLVIKGRFQQKQ 266
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHAV 117
IE +LR+YI EYVTC C+SP+T + +D RL+ C C + SV +IK+GF AV
Sbjct 267 IENVLRRYIKEYVTCHTCRSPDTILQKD--IRLYFLQCETCHSRCSVASIKTGFQAV 321
> 7294562
Length=312
Score = 140 bits (354), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 63/117 (53%), Positives = 86/117 (73%), Gaps = 2/117 (1%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
PQV+RVG+KK ++ NF DI +HR +H+L F+LAELGT GS+ G+ QL++KG++ PK
Sbjct 186 PQVLRVGTKKTSFANFMDIAKTLHRLPKHLLDFLLAELGTSGSMDGNQQLIIKGRFQPKQ 245
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHAV 117
IE +LR+YI EYVTC C+SP T + +D TRL+ C +CG+ SV +IKSGF AV
Sbjct 246 IENVLRRYIKEYVTCHTCRSPETILQKD--TRLFFLQCESCGSRCSVASIKSGFQAV 300
> At5g20920
Length=268
Score = 137 bits (344), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 59/116 (50%), Positives = 86/116 (74%), Gaps = 2/116 (1%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
PQV+R G+KK ++NF D+C MHR +HV+Q++LAELGT GS+ G +LV+KG++ PK+
Sbjct 148 PQVLREGTKKTVFVNFMDLCKTMHRQPDHVMQYLLAELGTSGSLDGQQRLVVKGRFAPKN 207
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHA 116
E +LR+YIT+YV C CKSP+T +++++ RL+ C CG+ RSV IK+GF A
Sbjct 208 FEGILRRYITDYVICLGCKSPDTILSKEN--RLFFLRCEKCGSQRSVAPIKTGFVA 261
> CE16227
Length=250
Score = 136 bits (342), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 66/117 (56%), Positives = 83/117 (70%), Gaps = 2/117 (1%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
P+V R GSKK A+ NF +IC +M R +HVLQF+LAELGT GSI G L++KG++ K
Sbjct 119 PEVARAGSKKTAFSNFLEICRLMKRQDKHVLQFLLAELGTTGSIDGSNCLIVKGRWQQKQ 178
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHAV 117
E++LRKYI EYV C CKSP T +T+D TRL+ C+ CG+ SVT IKSGF AV
Sbjct 179 FESVLRKYIKEYVMCHTCKSPETQLTKD--TRLFFLQCTNCGSRCSVTAIKSGFKAV 233
> YPL237w
Length=285
Score = 124 bits (312), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 58/120 (48%), Positives = 81/120 (67%), Gaps = 1/120 (0%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
P V KK + N +DI +HR EH++Q++ AELGT GS+ G +LV+KGK+ K
Sbjct 162 PPVCLRDGKKTIFSNIQDIAEKLHRSPEHLIQYLFAELGTSGSVDGQKRLVIKGKFQSKQ 221
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHA-VGK 119
+E +LR+YI EYVTC+ CKS NT + R+ RL+ C +CG+ RSV++IK+GF A VGK
Sbjct 222 MENVLRRYILEYVTCKTCKSINTELKREQSNRLFFMVCKSCGSTRSVSSIKTGFQATVGK 281
> SPAC6B12.17c
Length=310
Score = 124 bits (310), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 59/120 (49%), Positives = 84/120 (70%), Gaps = 4/120 (3%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
P V R G KK + N DI MHR +HV+QF+ AELGT GS+ G +L++KG++ K
Sbjct 189 PSVHREG-KKTIFANISDISKRMHRSLDHVIQFLFAELGTSGSVDGSSRLIIKGRFQQKQ 247
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHA-VGK 119
IE +LR+YI EYVTC+ CKSP+T +T+++ R++ +C ACG+ RSV IK+G+ A +GK
Sbjct 248 IENVLRRYIVEYVTCKTCKSPDTILTKEN--RIFFMTCEACGSVRSVQAIKTGYQAQIGK 305
> At3g07920
Length=169
Score = 88.6 bits (218), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 40/94 (42%), Positives = 61/94 (64%), Gaps = 4/94 (4%)
Query 1 PQVVRV----GSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKY 56
PQV+R G+KK ++NF D C MHR +HV+ F+LAELGT G + +LV++G++
Sbjct 46 PQVLREETAKGTKKTVFVNFMDYCKTMHRNPDHVMAFLLAELGTRGKLDDQQRLVVRGRF 105
Query 57 GPKHIEALLRKYITEYVTCQMCKSPNTSMTRDSR 90
+ E+LLR YI +YV C CKS +++++R
Sbjct 106 TQRDFESLLRGYILDYVMCIPCKSTEAILSKENR 139
> ECU06g0210
Length=228
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/120 (37%), Positives = 70/120 (58%), Gaps = 4/120 (3%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
P V+ ++K + IN +I I++R ++H++ F+ EL T GSI DG+L+LKG +
Sbjct 103 PLDVKREARKTS-INIAEISRILNRSADHLVSFLCNELMTTGSINKDGKLLLKGMFIRSE 161
Query 61 IEALLRKYITEYVTCQMCKS-PNTSMTRDSRTRLWHQSCSACGANRSVTTIKSGFHAVGK 119
I+ +LR+YI +V C++C+S +T + RD RL+ CS CG R V G GK
Sbjct 162 IQEVLRRYIEHFVVCKICESVEDTDIVRDK--RLYFLKCSKCGGARCVGNAVEGITTKGK 219
> At5g01940
Length=231
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 56/88 (63%), Gaps = 1/88 (1%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKH 60
PQ++ G+ V +NF D+C MHR +HV++F+LA++ T+ S+ +L +KG K
Sbjct 105 PQLLAEGTITVC-LNFADLCRTMHRKPDHVMKFLLAQMETKVSLNKQQRLEIKGLVSSKD 163
Query 61 IEALLRKYITEYVTCQMCKSPNTSMTRD 88
+A+ RKYI +V C CKSP+T++ +
Sbjct 164 FQAVFRKYIDAFVICVCCKSPDTALAEE 191
> Hs17476977
Length=115
Score = 67.0 bits (162), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 49/87 (56%), Gaps = 12/87 (13%)
Query 31 LQFVLAELGTEGSIAGDGQLVLKGKYGPKHIEALLRKYITEYVTCQMCKSPNTSMTRDSR 90
L F LAELGT GSI ++ E +LR+YI EYVTC C+SP+T + +D
Sbjct 29 LAFSLAELGTSGSIDDSN----------RNRENVLRRYIKEYVTCHTCQSPDTILQKD-- 76
Query 91 TRLWHQSCSACGANRSVTTIKSGFHAV 117
TRL+ C C + SV +IK+ F V
Sbjct 77 TRLYFLQCKTCHSRCSVASIKTRFQVV 103
> ECU07g1330
Length=281
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 54/110 (49%), Gaps = 4/110 (3%)
Query 2 QVVRVGSKKVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKHI 61
Q + G+ + +N ++I + + RP ++L+F+ EL T + G G+ + GKY I
Sbjct 24 QEGKAGNTRTLIVNLEEIGSSLKRPPLYILKFMSYELATRTDV-GKGRYAVNGKYESSRI 82
Query 62 EALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANRSVTTIK 111
+ L+ +I E+V C C +P T + C ACG ++ + K
Sbjct 83 QDLIYNFIDEFVICPFCSNPETFYINSGGLSM---ECLACGKRSNMRSSK 129
> YPR041w
Length=405
Score = 53.9 bits (128), Expect = 7e-08, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 55/96 (57%), Gaps = 6/96 (6%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGD-GQLVLKGKYGPKHIEALLRKY 68
K A +N DI ++RP+ +++++ ELG + SI+ D + ++ G + P ++ +L +
Sbjct 33 KTAVLNVADISHALNRPAPYIVKYFGFELGAQTSISVDKDRYLVNGVHEPAKLQDVLDGF 92
Query 69 ITEYVTCQMCKSPNTS--MTRDSRTRLWHQSCSACG 102
I ++V C CK+P T +T+D+ + C ACG
Sbjct 93 INKFVLCGSCKNPETEIIITKDNDLV---RDCKACG 125
> SPAC2F7.05c
Length=395
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 45/89 (50%), Gaps = 1/89 (1%)
Query 15 NFKDICAIMHRPSEHVLQFVLAELGTEGSIAGD-GQLVLKGKYGPKHIEALLRKYITEYV 73
N DI + RP +V +F ELG + +I D + ++ G + ++ LL +I +V
Sbjct 39 NMSDIAKALGRPPLYVTKFFGFELGAQTTIIADMDRYIVNGAHDAGKLQDLLDVFIRRFV 98
Query 74 TCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
C C++P T ++ + + + C ACG
Sbjct 99 LCASCQNPETELSINKKDQTISYDCKACG 127
> 7293169
Length=464
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 48/94 (51%), Gaps = 1/94 (1%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSIA-GDGQLVLKGKYGPKHIEALLRKY 68
K +N ++ + RP+ + ++ ELG + + + V+ G + ++ LL +
Sbjct 34 KTVLVNMAEVARAIGRPATYPTKYFGCELGAQTLFDHKNERFVVNGSHDVNKLQDLLDGF 93
Query 69 ITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
I ++V C C +P T++T ++ + QSC ACG
Sbjct 94 IRKFVLCPECDNPETNLTVSAKNQTISQSCKACG 127
> At1g36730
Length=439
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 2/94 (2%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSI-AGDGQLVLKGKYGPKHIEALLRKY 68
K +N +I + RP+ + ++ ELG + +G ++ G + + LL +
Sbjct 35 KTNVVNMVEIAKALGRPAAYTTKYFGCELGAQSKFDEKNGTSLVNGAHDTSKLAGLLENF 94
Query 69 ITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
I +YV C C +P T + ++T++ C+ACG
Sbjct 95 IKKYVQCYGCGNPETEILI-TKTQMLQLKCAACG 127
> HsM4503543
Length=431
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELG--TEGSIAGDGQLVLKGKYGPKHIEALLRK 67
K +N D+ ++RP + ++ ELG T+ + D + ++ G + ++ +L
Sbjct 33 KTVIVNMVDVAKALNRPPTYPTKYFGCELGAQTQFDVKND-RYIVNGSHEANKLQDMLDG 91
Query 68 YITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
+I ++V C C++P T + + + + SC ACG
Sbjct 92 FIKKFVLCPECENPETDLHVNPKKQTIGNSCKACG 126
> Hs21361337
Length=431
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELG--TEGSIAGDGQLVLKGKYGPKHIEALLRK 67
K +N D+ ++RP + ++ ELG T+ + D + ++ G + ++ +L
Sbjct 33 KTVIVNMVDVAKALNRPPTYPTKYFGCELGAQTQFDVKND-RYIVNGSHEANKLQDMLDG 91
Query 68 YITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
+I ++V C C++P T + + + + SC ACG
Sbjct 92 FIKKFVLCPECENPETDLHVNPKKQTIGNSCKACG 126
> At1g77840
Length=437
Score = 43.9 bits (102), Expect = 7e-05, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 45/94 (47%), Gaps = 2/94 (2%)
Query 10 KVAWINFKDICAIMHRPSEHVLQFVLAELGTEGSI-AGDGQLVLKGKYGPKHIEALLRKY 68
K IN +I + RP + ++ ELG + G ++ G + + LL +
Sbjct 35 KTNIINNVEIAKALARPPSYTTKYFGCELGAQSKFDEKTGTSLVNGAHNTSKLAGLLENF 94
Query 69 ITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACG 102
I ++V C C +P T + ++T++ + C+ACG
Sbjct 95 IKKFVQCYGCGNPETEIII-TKTQMVNLKCAACG 127
> CE27367
Length=436
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/92 (22%), Positives = 47/92 (51%), Gaps = 5/92 (5%)
Query 15 NFKDICAIMHRPSEHVLQFVLAELGTEGSI-AGDGQLVLKGKYGPKHIEALLRKYITEYV 73
N +I + RP + ++ ELG + + A + + ++ G++ ++ +L +I ++V
Sbjct 38 NMSEIAKALERPPMYPTKYFGCELGAQTNFDAKNERYIVNGEHDANKLQDILDGFIKKFV 97
Query 74 TCQMCKSPNTSM-TRDSRTRLWHQSCSACGAN 104
C+ C++P T + R + + C ACG +
Sbjct 98 LCKSCENPETQLFVRKNNIK---SKCKACGCS 126
> Hs22042204
Length=199
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 29 HVLQFVLAELGTEGSIAGDGQLVLKGKYGPKHIEALLR 66
H+ F+L ELGT GS+ G+ QLV+KG+ K I+ +L
Sbjct 73 HLPAFLLGELGTNGSVGGNNQLVIKGRLQKKQIKIVLE 110
> CE24496
Length=170
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 35/81 (43%), Gaps = 4/81 (4%)
Query 19 ICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPKHIEALLRKYITEYVTCQMC 78
CAI+ S + +VLA+ + IA G+L++ P L +TE CQ+C
Sbjct 6 FCAILAVISAISVSYVLAQRSQQSVIADLGKLIVDN-CPPMLCTGLDCAVVTERNGCQLC 64
Query 79 KSPNTSMTRDSRTR---LWHQ 96
P S +R LWH
Sbjct 65 ACPIGSPSRGCDPMPFILWHD 85
> Hs17479759
Length=129
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 13 WINFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGK 55
+I+ ++C +M P E + + E+ E I GDGQ +L G+
Sbjct 48 YISAAELCHVMTNPGEKLTDDKVDEMIREAGIDGDGQDLLFGE 90
> Hs21361124
Length=279
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 21/106 (19%)
Query 2 QVVRVGSKKVAW--INFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPK 59
+ + G++K+ + ++ + C I HR + + GT+ I D Q Y +
Sbjct 106 KTIMPGTRKMEYKGSSWHETCFICHRCQQPI--------GTKSFIPKDNQNFCVPCYEKQ 157
Query 60 HIEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANR 105
H + C CK P T+ R + WH+ C C A R
Sbjct 158 HA-----------MQCVQCKKPITTGGVTYREQPWHKECFVCTACR 192
> HsM4503723
Length=279
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 21/106 (19%)
Query 2 QVVRVGSKKVAW--INFKDICAIMHRPSEHVLQFVLAELGTEGSIAGDGQLVLKGKYGPK 59
+ + G++K+ + ++ + C I HR + + GT+ I D Q Y +
Sbjct 106 KTIMPGTRKMEYKGSSWHETCFICHRCQQPI--------GTKSFIPKDNQNFCVPCYEKQ 157
Query 60 HIEALLRKYITEYVTCQMCKSPNTSMTRDSRTRLWHQSCSACGANR 105
H + C CK P T+ R + WH+ C C A R
Sbjct 158 HA-----------MQCVQCKMPITTGGVTYREQPWHKECFVCTACR 192
> CE28943
Length=476
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 47 DGQLVLKGKYGPKHIEALLRKYITEYVTCQMCKSPNTSMTRDSRTRL 93
+ +++L Y P+H+ +L T ++C NT MTRD+ T L
Sbjct 226 EDKIILLKNYAPQHL--ILMPAFRSPDTTRVCLFNNTYMTRDNNTDL 270
> CE12084
Length=477
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 47 DGQLVLKGKYGPKHIEALLRKYITEYVTCQMCKSPNTSMTRDSRTRL 93
+ +++L Y P+H+ +L T ++C NT MTRD+ T L
Sbjct 227 EDKIILLKNYAPQHL--ILMPAFRSPDTTRVCLFNNTYMTRDNNTDL 271
> SPBC244.03
Length=796
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 8/48 (16%)
Query 1 PQVVRVGSKKVAWINFKDICAIMHRPSEH--------VLQFVLAELGT 40
P V S KV W N+ ++ + M RP++ ++ + ELGT
Sbjct 303 PSVTMDSSGKVVWSNYNEVMSAMIRPAKEQSDLTDGSLISLSVKELGT 350
Lambda K H
0.321 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40