bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2849_orf2
Length=138
Score E
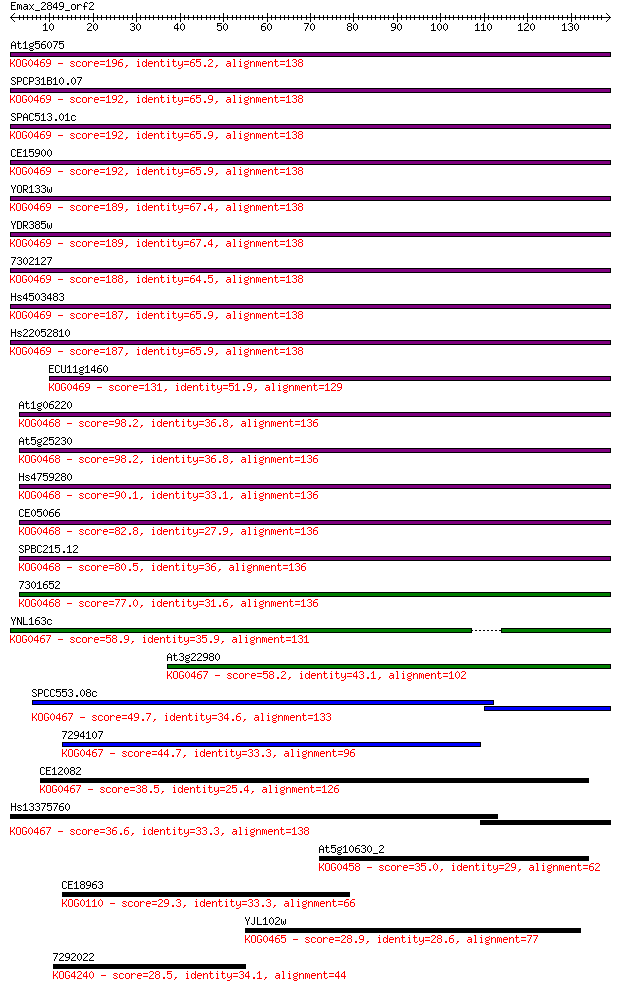
Sequences producing significant alignments: (Bits) Value
At1g56075 196 1e-50
SPCP31B10.07 192 2e-49
SPAC513.01c 192 2e-49
CE15900 192 2e-49
YOR133w 189 2e-48
YDR385w 189 2e-48
7302127 188 2e-48
Hs4503483 187 4e-48
Hs22052810 187 5e-48
ECU11g1460 131 4e-31
At1g06220 98.2 4e-21
At5g25230 98.2 4e-21
Hs4759280 90.1 1e-18
CE05066 82.8 2e-16
SPBC215.12 80.5 9e-16
7301652 77.0 9e-15
YNL163c 58.9 3e-09
At3g22980 58.2 5e-09
SPCC553.08c 49.7 1e-06
7294107 44.7 6e-05
CE12082 38.5 0.004
Hs13375760 36.6 0.016
At5g10630_2 35.0 0.039
CE18963 29.3 2.2
YJL102w 28.9 3.4
7292022 28.5 4.2
> At1g56075
Length=855
Score = 196 bits (498), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 90/138 (65%), Positives = 108/138 (78%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I ++ +T MNDQKDK ML LG+ +K D+K+L GK L+KRVMQ WLPA LEM++
Sbjct 291 IKQIIATCMNDQKDKLWPMLAKLGVSMKNDEKELMGKPLMKRVMQTWLPASTALLEMMIF 350
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQ+YRV+ LYEGP DD+ AN IRNCDPN PLM+YVSKM+P SDKGRF+AFGRV
Sbjct 351 HLPSPHTAQRYRVENLYEGPLDDQYANAIRNCDPNGPLMLYVSKMIPASDKGRFFAFGRV 410
Query 121 FSGTVATGQKVRIQGPYY 138
F+G V+TG KVRI GP Y
Sbjct 411 FAGKVSTGMKVRIMGPNY 428
> SPCP31B10.07
Length=842
Score = 192 bits (488), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 91/138 (65%), Positives = 110/138 (79%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I ++F +MN +KD+ +L+ L + +K D+K+L GKALLK VM+ +LPA D +EMIV
Sbjct 281 IYRIFDAVMNSRKDEVFTLLSKLEVTIKPDEKELEGKALLKVVMRKFLPAADALMEMIVL 340
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQ+YR +TLYEGP DDE A GIRNCD NAPLM+YVSKMVPTSD+GRFYAFGRV
Sbjct 341 HLPSPKTAQQYRAETLYEGPMDDECAVGIRNCDANAPLMIYVSKMVPTSDRGRFYAFGRV 400
Query 121 FSGTVATGQKVRIQGPYY 138
FSGTV +G KVRIQGP Y
Sbjct 401 FSGTVRSGLKVRIQGPNY 418
> SPAC513.01c
Length=812
Score = 192 bits (488), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 91/138 (65%), Positives = 110/138 (79%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I ++F +MN +KD+ +L+ L + +K D+K+L GKALLK VM+ +LPA D +EMIV
Sbjct 251 IYRIFDAVMNSRKDEVFTLLSKLEVTIKPDEKELEGKALLKVVMRKFLPAADALMEMIVL 310
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQ+YR +TLYEGP DDE A GIRNCD NAPLM+YVSKMVPTSD+GRFYAFGRV
Sbjct 311 HLPSPKTAQQYRAETLYEGPMDDECAVGIRNCDANAPLMIYVSKMVPTSDRGRFYAFGRV 370
Query 121 FSGTVATGQKVRIQGPYY 138
FSGTV +G KVRIQGP Y
Sbjct 371 FSGTVRSGLKVRIQGPNY 388
> CE15900
Length=852
Score = 192 bits (487), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 91/138 (65%), Positives = 105/138 (76%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I +F +MN +KDK ++ LGI+L D+KDL GK L+K M+ WLPAGD L+MI
Sbjct 291 IFMVFDAVMNIKKDKTAALVEKLGIKLANDEKDLEGKPLMKVFMRKWLPAGDTMLQMIAF 350
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQKYR++ LYEGP DDEAA I+ CDPN PLMMY+SKMVPTSDKGRFYAFGRV
Sbjct 351 HLPSPVTAQKYRMEMLYEGPHDDEAAVAIKTCDPNGPLMMYISKMVPTSDKGRFYAFGRV 410
Query 121 FSGTVATGQKVRIQGPYY 138
FSG VATG K RIQGP Y
Sbjct 411 FSGKVATGMKARIQGPNY 428
> YOR133w
Length=842
Score = 189 bits (479), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 93/138 (67%), Positives = 106/138 (76%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I +LF+ IMN +KD+ +L L I LKGD+KDL GKALLK VM+ +LPA D LEMIV
Sbjct 281 IFRLFTAIMNFKKDEIPVLLEKLEIVLKGDEKDLEGKALLKVVMRKFLPAADALLEMIVL 340
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQ YR + LYEGP DD I+NCDP A LM+YVSKMVPTSDKGRFYAFGRV
Sbjct 341 HLPSPVTAQAYRAEQLYEGPADDANCIAIKNCDPKADLMLYVSKMVPTSDKGRFYAFGRV 400
Query 121 FSGTVATGQKVRIQGPYY 138
F+GTV +GQKVRIQGP Y
Sbjct 401 FAGTVKSGQKVRIQGPNY 418
> YDR385w
Length=842
Score = 189 bits (479), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 93/138 (67%), Positives = 106/138 (76%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I +LF+ IMN +KD+ +L L I LKGD+KDL GKALLK VM+ +LPA D LEMIV
Sbjct 281 IFRLFTAIMNFKKDEIPVLLEKLEIVLKGDEKDLEGKALLKVVMRKFLPAADALLEMIVL 340
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQ YR + LYEGP DD I+NCDP A LM+YVSKMVPTSDKGRFYAFGRV
Sbjct 341 HLPSPVTAQAYRAEQLYEGPADDANCIAIKNCDPKADLMLYVSKMVPTSDKGRFYAFGRV 400
Query 121 FSGTVATGQKVRIQGPYY 138
F+GTV +GQKVRIQGP Y
Sbjct 401 FAGTVKSGQKVRIQGPNY 418
> 7302127
Length=832
Score = 188 bits (478), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 89/138 (64%), Positives = 108/138 (78%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I K+F IMN +K++ +L +G+ LK +DKD GKALLK VM+ WLPAG+ L+MI
Sbjct 271 IYKVFDAIMNYKKEEIGTLLEKIGVTLKHEDKDKDGKALLKTVMRTWLPAGEALLQMIAI 330
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQKYR++ LYEGP DDEAA +++CDP+ PLMMY+SKMVPTSDKGRFYAFGRV
Sbjct 331 HLPSPVVAQKYRMEMLYEGPHDDEAAIAVKSCDPDGPLMMYISKMVPTSDKGRFYAFGRV 390
Query 121 FSGTVATGQKVRIQGPYY 138
F+G VATGQK RI GP Y
Sbjct 391 FAGKVATGQKCRIMGPNY 408
> Hs4503483
Length=858
Score = 187 bits (476), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 91/138 (65%), Positives = 106/138 (76%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I K+F IMN +K++ K++ L I+L +DKD GK LLK VM+ WLPAGD L+MI
Sbjct 297 IFKVFDAIMNFKKEETAKLIEKLDIKLDSEDKDKEGKPLLKAVMRRWLPAGDALLQMITI 356
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQKYR + LYEGP DDEAA GI++CDP PLMMY+SKMVPTSDKGRFYAFGRV
Sbjct 357 HLPSPVTAQKYRCELLYEGPPDDEAAMGIKSCDPKGPLMMYISKMVPTSDKGRFYAFGRV 416
Query 121 FSGTVATGQKVRIQGPYY 138
FSG V+TG KVRI GP Y
Sbjct 417 FSGLVSTGLKVRIMGPNY 434
> Hs22052810
Length=846
Score = 187 bits (475), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 91/138 (65%), Positives = 106/138 (76%), Gaps = 0/138 (0%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR 60
I K+F IMN +K++ K++ L I+L +DKD GK LLK VM+ WLPAGD L+MI
Sbjct 285 IFKVFDAIMNFKKEETAKLIEKLDIKLDSEDKDKEGKPLLKAVMRRWLPAGDALLQMITI 344
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
HLPSP AQKYR + LYEGP DDEAA GI++CDP PLMMY+SKMVPTSDKGRFYAFGRV
Sbjct 345 HLPSPVTAQKYRCELLYEGPPDDEAAMGIKSCDPKGPLMMYISKMVPTSDKGRFYAFGRV 404
Query 121 FSGTVATGQKVRIQGPYY 138
FSG V+TG KVRI GP Y
Sbjct 405 FSGLVSTGLKVRIMGPNY 422
> ECU11g1460
Length=850
Score = 131 bits (330), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 67/131 (51%), Positives = 87/131 (66%), Gaps = 3/131 (2%)
Query 10 NDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHLPSPWEAQ 69
N + ++ ++ L ++ KG SGK+L K VM+ WLPA DC LE I LPSP ++Q
Sbjct 295 NGKVEEIKEYLKFYKVDFKGVVLTGSGKSLFKEVMKTWLPAADCILEQIALKLPSPLQSQ 354
Query 70 KYRVDTLYEGPKDDEAANGIRNCDPN--APLMMYVSKMVPTSDKGRFYAFGRVFSGTVAT 127
K R D LYEGP DDE AN I+ CD + AP+ MYVSKM+P++D RF AFGRVFSG +
Sbjct 355 KLRYDYLYEGPADDEVANAIKMCDGSDEAPVSMYVSKMIPSND-NRFIAFGRVFSGKIFP 413
Query 128 GQKVRIQGPYY 138
G K+R+Q P Y
Sbjct 414 GMKIRVQEPGY 424
> At1g06220
Length=987
Score = 98.2 bits (243), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 50/136 (36%), Positives = 73/136 (53%), Gaps = 0/136 (0%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHL 62
K++S ++ + K E L LG+ L L+ + LL+ + +M+V+H+
Sbjct 393 KIYSQVIGEHKKSVETTLAELGVTLSNSAYKLNVRPLLRLACSSVFGSASGFTDMLVKHI 452
Query 63 PSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRVFS 122
PSP EA +VD Y G KD + CDP+ PLM+ V+K+ P SD F FGRV+S
Sbjct 453 PSPREAAARKVDHSYTGTKDSPIYESMVECDPSGPLMVNVTKLYPKSDTSVFDVFGRVYS 512
Query 123 GTVATGQKVRIQGPYY 138
G + TGQ VR+ G Y
Sbjct 513 GRLQTGQSVRVLGEGY 528
> At5g25230
Length=973
Score = 98.2 bits (243), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 50/136 (36%), Positives = 73/136 (53%), Gaps = 0/136 (0%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHL 62
K++S ++ + K E L LG+ L L+ + LL+ + +M+V+H+
Sbjct 379 KIYSQVIGEHKKSVETTLAELGVTLSNSAYKLNVRPLLRLACSSVFGSASGFTDMLVKHI 438
Query 63 PSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRVFS 122
PSP EA +VD Y G KD + CDP+ PLM+ V+K+ P SD F FGRV+S
Sbjct 439 PSPREAAARKVDHSYTGTKDSPIYESMVECDPSGPLMVNVTKLYPKSDTSVFDVFGRVYS 498
Query 123 GTVATGQKVRIQGPYY 138
G + TGQ VR+ G Y
Sbjct 499 GRLQTGQSVRVLGEGY 514
> Hs4759280
Length=972
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/136 (33%), Positives = 75/136 (55%), Gaps = 0/136 (0%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHL 62
K+ + ++ D + L LGI L ++ L+ + LL+ V + + ++M V+H+
Sbjct 379 KILAQVVGDVDTSLPRTLDELGIHLTKEELKLNIRPLLRLVCKKFFGEFTGFVDMCVQHI 438
Query 63 PSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRVFS 122
PSP K +++ Y G D + + +CDP+ PLM + +KM T D +F+AFGRV S
Sbjct 439 PSPKVGAKPKIEHTYTGGVDSDLGEAMSDCDPDGPLMCHTTKMYSTDDGVQFHAFGRVLS 498
Query 123 GTVATGQKVRIQGPYY 138
GT+ GQ V++ G Y
Sbjct 499 GTIHAGQPVKVLGENY 514
> CE05066
Length=974
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/136 (27%), Positives = 74/136 (54%), Gaps = 0/136 (0%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHL 62
K+FS ++ D ++ LGI L +++ ++ + L+ + + + ++++V+++
Sbjct 383 KIFSQVVGDVDTCLPDVMAELGIRLSKEEQKMNVRPLIALICKRFFGDFSAFVDLVVQNI 442
Query 63 PSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRVFS 122
SP E K +++ Y GP D + A ++ C+ PLM++ +K P D +F+ FGRV S
Sbjct 443 KSPLENAKTKIEQTYLGPADSQLAQEMQKCNAEGPLMVHTTKNYPVDDATQFHVFGRVMS 502
Query 123 GTVATGQKVRIQGPYY 138
GT+ VR+ G Y
Sbjct 503 GTLEANTDVRVLGENY 518
> SPBC215.12
Length=983
Score = 80.5 bits (197), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 74/140 (52%), Gaps = 6/140 (4%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWL--PAGDCPLEMIVR 60
KL + ++D+ +K +K L++ I LK D L K LL+ + + P G + + R
Sbjct 390 KLHTLTISDEAEKLKKHLSSFQIYLKPKDYLLDPKPLLQLICASFFGFPVGF--VNAVTR 447
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNC--DPNAPLMMYVSKMVPTSDKGRFYAFG 118
H+PSP E + Y GP + I + +APL+M+V+K+ T D FYAF
Sbjct 448 HIPSPRENAARKASQSYIGPINSSIGKAILEMSREESAPLVMHVTKLYNTVDANNFYAFA 507
Query 119 RVFSGTVATGQKVRIQGPYY 138
RV+SG V GQKV++ G Y
Sbjct 508 RVYSGQVKKGQKVKVLGENY 527
> 7301652
Length=975
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 71/138 (51%), Gaps = 4/138 (2%)
Query 3 KLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCP--LEMIVR 60
KL + ++ D L L + + ++ + + LL+ V ++ GDC ++M V
Sbjct 382 KLIAQVVGDVDTTLSDTLAELNVRVSKEEMKSNIRPLLRLVCNRFM--GDCSGFVDMCVE 439
Query 61 HLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRFYAFGRV 120
H+ SP E K +VD +Y GPK+ + + +C+ LM++ SKM P D F R+
Sbjct 440 HIKSPLENAKRKVDHIYTGPKEGDIYRDMISCNQYGTLMVHSSKMYPNDDCTFFQVLARI 499
Query 121 FSGTVATGQKVRIQGPYY 138
SGT+ GQ+VR+ G Y
Sbjct 500 VSGTLHAGQEVRVLGENY 517
> YNL163c
Length=1110
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 57/113 (50%), Gaps = 7/113 (6%)
Query 1 ICKLFSTIMNDQ-KDKYEKMLTTLGIELKGDD-KDLSGKALLKRVMQLWLPAGDCPLEMI 58
I K++ I+ + + EK+ TL I+L D + K LL+ +M WLP L +
Sbjct 302 IWKIYQNIITSRDSEMVEKIAKTLNIKLLARDLRSKDDKQLLRTIMGQWLPVSTAVLLTV 361
Query 59 VRHLPSPWEAQKYRVDTLYEGPKDDEAAN-----GIRNCDPNAPLMMYVSKMV 106
+ LPSP E+Q R++T+ D A + ++ CD P+ YVSKM+
Sbjct 362 IEKLPSPLESQTDRLNTILVSESDTAAMDPRLLKAMKTCDKEGPVSAYVSKML 414
Score = 32.7 bits (73), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 114 FYAFGRVFSGTVATGQKVRIQGPYY 138
AF R++SGT+ GQ++ + GP Y
Sbjct 582 LVAFARIYSGTLRVGQEISVLGPKY 606
> At3g22980
Length=1015
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 60/143 (41%), Gaps = 41/143 (28%)
Query 37 KALLKRVMQLWLPAGDCPLEMIVRHLPSPWEAQKYRVDTLYEGPK----DDEAAN----- 87
K +L+ VM WLP D L M V+HLP P AQ YR+ L K DD ++
Sbjct 322 KNVLQSVMSRWLPLSDAVLSMAVKHLPDPIAAQAYRIPRLVPERKIIGGDDVDSSVLAEA 381
Query 88 -----GIRNCD--PNAPLMMYVS-------KMVPTSDKGR------------------FY 115
I CD ++P +++VS KM+P R F
Sbjct 382 ELVRKSIEACDSSSDSPCVVFVSKMFAIPMKMIPQDGNHRERMNGLNDDDSKSESDECFL 441
Query 116 AFGRVFSGTVATGQKVRIQGPYY 138
AF R+FSG + GQ+V + Y
Sbjct 442 AFARIFSGVLRAGQRVFVITALY 464
> SPCC553.08c
Length=1000
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/114 (30%), Positives = 54/114 (47%), Gaps = 8/114 (7%)
Query 6 STIMNDQKDKYEKMLTTLGIE-LKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHLPS 64
S + N + EK++ L I+ L D K + LL + Q WLP L +R +PS
Sbjct 294 SAVSNRNLENIEKIIKALNIKVLPRDIKSKDPRNLLLAIFQQWLPLSTAILLTAIREIPS 353
Query 65 PWEAQKYRV-DTLYEGPK----DDEAANGIRNCDPN--APLMMYVSKMVPTSDK 111
P AQ R L P D + + +CD + P+++Y+SKMV S++
Sbjct 354 PINAQANRARKVLSSTPHYEMIDPDITLAMESCDASKEQPVLVYISKMVAFSER 407
Score = 35.0 bits (79), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 110 DKGRFYAFGRVFSGTVATGQKVRIQGPYY 138
DK F R++SGT++ GQ+V + GP Y
Sbjct 464 DKDILIGFARIYSGTISVGQEVYVYGPKY 492
> 7294107
Length=860
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 49/107 (45%), Gaps = 11/107 (10%)
Query 13 KDKYEKMLTTLGIELKGDDKDLSGKAL-LKRVMQLWLPAGDCPLEMIVRHLPSPWEAQKY 71
KDK + LG++L D L+ L +K V+ WLP L M+++H+P P +
Sbjct 284 KDKLPGIAEKLGLKLATRDLRLTDPKLQIKAVLGQWLPIDKSVLHMVIQHVPPPHKISDE 343
Query 72 R---------VDTLYEGPKDDEAANGIRNCDPNAP-LMMYVSKMVPT 108
R VD P+ E +CD N+ ++ +VSKM P
Sbjct 344 RAQRLLYPANVDLSSLPPETLELKESFTSCDANSSNVIAFVSKMTPV 390
> CE12082
Length=906
Score = 38.5 bits (88), Expect = 0.004, Method: Composition-based stats.
Identities = 32/130 (24%), Positives = 57/130 (43%), Gaps = 9/130 (6%)
Query 8 IMNDQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVR--HLPSP 65
++ + K + LGI LK + + +M+ WLP + R ++ S
Sbjct 283 LVENDATKLAEAAKKLGINLKSRRANEA----FDELMRTWLPLPKASFRAVARAPNVRST 338
Query 66 WEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKG--RFYAFGRVFSG 123
++ Q +R+D L D + C+P A +++V K++ T DK A RV SG
Sbjct 339 FDTQ-HRLDHLTGHRLDHPLRKFVLECNPEAMTLVFVVKLLQTEDKNLTSRRAICRVLSG 397
Query 124 TVATGQKVRI 133
T+ G + +
Sbjct 398 TLRKGDTLYV 407
> Hs13375760
Length=857
Score = 36.6 bits (83), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 57/124 (45%), Gaps = 12/124 (9%)
Query 1 ICKLFSTIMNDQKDKYEKMLTTLGIELKGDDKDLSG-KALLKRVMQLWLPAGDCPLEMIV 59
I L+ ++ KDK +K++T+LG+++ + S K + + WLP L M+
Sbjct 38 IWSLYDAVLKKDKDKIDKIVTSLGLKIGAREARHSDPKVQINAICSQWLPISHAVLAMVC 97
Query 60 RHLPSPWEAQKYRVD--------TLYEGPKDDEAAN-GIRNC--DPNAPLMMYVSKMVPT 108
+ LPSP + RV+ T P + +A C + AP++++VSKM
Sbjct 98 QKLPSPLDITAERVERLMCTGSQTFDSFPPETQALKAAFMKCGSEDTAPVIIFVSKMFAV 157
Query 109 SDKG 112
K
Sbjct 158 DAKA 161
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 109 SDKGRFYAFGRVFSGTVATGQKVRIQGPYY 138
+++ F AF RVFSG G+K+ + GP Y
Sbjct 237 NNQESFIAFARVFSGVARRGKKIFVLGPKY 266
> At5g10630_2
Length=462
Score = 35.0 bits (79), Expect = 0.039, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 37/64 (57%), Gaps = 2/64 (3%)
Query 72 RVDTLYEGPKDDEAANGIRNCDPNA--PLMMYVSKMVPTSDKGRFYAFGRVFSGTVATGQ 129
R+ + Y+GP +A + +++ D + PL+M + V ++ +G+ A G++ +G V G
Sbjct 235 RLSSWYQGPCLLDAVDSVKSPDRDVSKPLLMPICDAVRSTSQGQVSACGKLEAGAVRPGS 294
Query 130 KVRI 133
KV +
Sbjct 295 KVMV 298
> CE18963
Length=872
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Query 13 KDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCPLEMIVRHLPSPWEAQKYR 72
KD ++L +ELK ++ + K LKR G+C +++VR+L P+EA
Sbjct 707 KDMQGELLDGHSLELKISHRENADKGALKRKEVKQKEQGECT-KLLVRNL--PFEASVKE 763
Query 73 VDTLYE 78
V+TL+E
Sbjct 764 VETLFE 769
> YJL102w
Length=819
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 55 LEMIVRHLPSPWEAQKYRVDTLYEGPKDDEAANGIRNCDPNAPLMMYVSKMVPTSDKGRF 114
L+ IV +LPSP EA+ ++ K D + N + N + + K++ +G+
Sbjct 316 LDAIVNYLPSPIEAELPELNDKTVPMKYDPKVGCLVNNNKNLCIALAF-KVITDPIRGK- 373
Query 115 YAFGRVFSGTVATGQKV 131
F R++SGT+ +G V
Sbjct 374 QIFIRIYSGTLNSGNTV 390
> 7292022
Length=2056
Score = 28.5 bits (62), Expect = 4.2, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 22/44 (50%), Gaps = 4/44 (9%)
Query 11 DQKDKYEKMLTTLGIELKGDDKDLSGKALLKRVMQLWLPAGDCP 54
D ++ K +T L IE D D+ GK LL ++ + P G P
Sbjct 204 DHHNEMRKKITKLPIE----DLDMQGKKLLAKINAMAPPGGQAP 243
Lambda K H
0.320 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40