bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2747_orf1
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
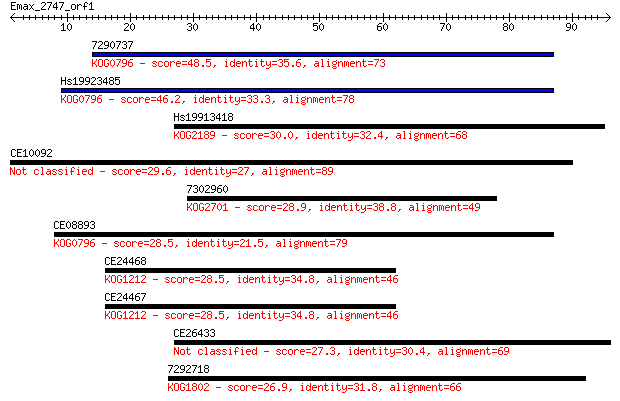
7290737 48.5 3e-06
Hs19923485 46.2 2e-05
Hs19913418 30.0 1.1
CE10092 29.6 1.6
7302960 28.9 2.3
CE08893 28.5 2.8
CE24468 28.5 3.0
CE24467 28.5 3.1
CE26433 27.3 6.7
7292718 26.9 9.6
> 7290737
Length=414
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 47/78 (60%), Gaps = 6/78 (7%)
Query 14 FEQEFARYLQRLVDQMENRIKRVQQRI-----DASNQPVELSKENEERVNAMNAEISELL 68
+E EF R+ ++ ++ +I++ +QR+ D N P LS+ E+ N + A I++LL
Sbjct 83 YEDEFLRFCNVMLHDVDRKIQKGKQRLLLMQRDQPNVPAPLSRHQEQLAN-LTARINKLL 141
Query 69 KQQDEAGSKGDIDAAEKL 86
+ +EAG +GD+D A+ L
Sbjct 142 SEAEEAGIRGDVDQAQDL 159
> Hs19923485
Length=432
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 49/83 (59%), Gaps = 5/83 (6%)
Query 9 FYQALFEQEFARYLQRLVDQMENRIKRVQQRI-----DASNQPVELSKENEERVNAMNAE 63
F + +E++F RYLQ L+ ++E RI+R R+ S+ + +NEE++ + +
Sbjct 76 FMKVGYERDFLRYLQSLLAEVERRIRRGHARLALSQNQQSSGAAGPTGKNEEKIQVLTDK 135
Query 64 ISELLKQQDEAGSKGDIDAAEKL 86
I LL+Q +E GS+G ++ A+ +
Sbjct 136 IDVLLQQIEELGSEGKVEEAQGM 158
> Hs19913418
Length=831
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 4/71 (5%)
Query 27 DQMENRIKRVQQRIDASNQPV-ELSKENEERVNAMNAEISEL--LKQQDEAGSKGDIDAA 83
DQ++NR+K++ + AS P E +E +E + +N I +L + Q E + + AA
Sbjct 227 DQLKNRVKKICEGFRASLYPCPETPQERKEMASGVNTRIDDLQMVLNQTEDHRQRVLQAA 286
Query 84 EKLNEKVAFLQ 94
K N +V F++
Sbjct 287 AK-NIRVWFIK 296
> CE10092
Length=5105
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 45/97 (46%), Gaps = 8/97 (8%)
Query 1 FEADPEREFYQALFEQEFARYLQRLVDQMENRIKRVQ----QRIDASNQPVELSKENEER 56
E RE +A+FE+ + Q ++++ E + + + D E E+E +
Sbjct 1395 LEMKNRRELQRAIFEKRIITFRQPVIEETEQELAIEENLNVEEDDLRTTSEESEGEDEVK 1454
Query 57 VNAMNAEISELLK----QQDEAGSKGDIDAAEKLNEK 89
++ +IS+ K +DEAG GD D+ E L ++
Sbjct 1455 NDSDPNDISDFWKIFKVTEDEAGVNGDRDSDEDLVQR 1491
> 7302960
Length=595
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 4/53 (7%)
Query 29 MENRIKRV----QQRIDASNQPVELSKENEERVNAMNAEISELLKQQDEAGSK 77
ME++I+R +Q DA N+ E +KE E+++N +N E+ QD+A K
Sbjct 310 MEHKIRRYCKENEQLQDAENEQKEKTKEVEDQLNTLNTELKAHKDTQDKADPK 362
> CE08893
Length=369
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/84 (20%), Positives = 47/84 (55%), Gaps = 5/84 (5%)
Query 8 EFYQALFEQEFARYLQRLVDQMENRIKRVQQRIDASNQPVELSKENEER-----VNAMNA 62
E+ Q FE+ R+L +L + + RI++ + ++ + + E E+R +N+++
Sbjct 77 EYGQLGFERRLMRFLVQLDEDNQRRIRKNKDKLSGMDDSGKRKLEEEKRQVQLEINSIDD 136
Query 63 EISELLKQQDEAGSKGDIDAAEKL 86
+ +++ ++AGS+G++ +++
Sbjct 137 VLKNMIRDAEQAGSEGNVTKCQEI 160
> CE24468
Length=615
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 9/55 (16%)
Query 16 QEFARYLQRLVDQMENRIKRVQQRIDASNQPVEL---------SKENEERVNAMN 61
+E + RLV++ ++R+Q R+ AS P+++ S E+ ERVN ++
Sbjct 561 EEMCLRVMRLVEESAEGVQRLQWRVGASAAPIDVENTPAGVVSSLEHFERVNLLH 615
> CE24467
Length=647
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 9/55 (16%)
Query 16 QEFARYLQRLVDQMENRIKRVQQRIDASNQPVEL---------SKENEERVNAMN 61
+E + RLV++ ++R+Q R+ AS P+++ S E+ ERVN ++
Sbjct 593 EEMCLRVMRLVEESAEGVQRLQWRVGASAAPIDVENTPAGVVSSLEHFERVNLLH 647
> CE26433
Length=613
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query 27 DQMENRIKRVQQRIDASNQPVELSKENEERVNAMNAEISELLKQQDEAGSKGDIDAAEK- 85
++ E R V +R+ + KE++E M EI ++++++ + KGDI A EK
Sbjct 299 EKSEARRITVNRRLRSGELEKNEQKEHKELRVKMREEIDKVIEEECKNKPKGDISAGEKP 358
Query 86 -----LNEKVAFLQR 95
LN F R
Sbjct 359 KQARLLNSSAEFFDR 373
> 7292718
Length=685
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 34/75 (45%), Gaps = 9/75 (12%)
Query 26 VDQMENRIKRVQQRI--------DASNQPVE-LSKENEERVNAMNAEISELLKQQDEAGS 76
VDQ+ +I R ++ +A + PV L+ N+ R N+E+ +L + +DE G
Sbjct 508 VDQLTEKIHRTNLKVVRVCAKSREAIDSPVSFLALHNQIRNMETNSELKKLQQLKDETGE 567
Query 77 KGDIDAAEKLNEKVA 91
D N K A
Sbjct 568 LSSADEKRYRNLKRA 582
Lambda K H
0.312 0.128 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40