bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
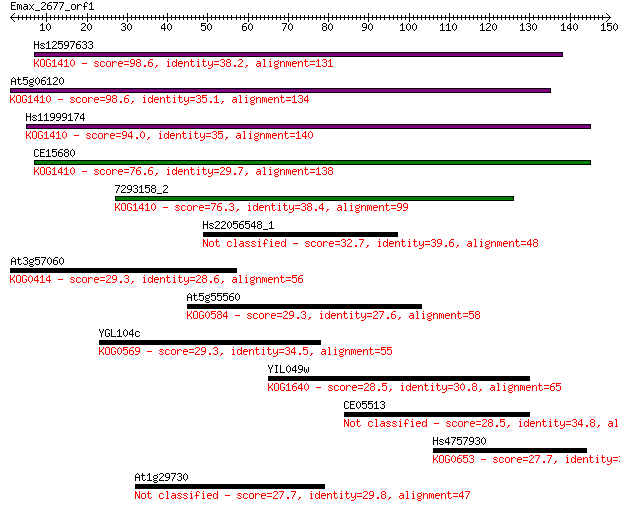
Query= Emax_2677_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
Hs12597633 98.6 4e-21
At5g06120 98.6 4e-21
Hs11999174 94.0 1e-19
CE15680 76.6 2e-14
7293158_2 76.3 2e-14
Hs22056548_1 32.7 0.28
At3g57060 29.3 2.9
At5g55560 29.3 3.0
YGL104c 29.3 3.1
YIL049w 28.5 4.4
CE05513 28.5 5.2
Hs4757930 27.7 7.9
At1g29730 27.7 8.4
> Hs12597633
Length=1088
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 78/132 (59%), Gaps = 1/132 (0%)
Query 7 RNKYMSAVILGAFKIIKTRTGLTDDLCYHEFCRLIGKINTSHHLSELCASEPFREFPQYL 66
R KY+ +I G +I++ GL+D YHEFCR + ++ T++ L EL + + E + +
Sbjct 299 RAKYLGNLIKGVKRILENPQGLSDPGNYHEFCRFLARLKTNYQLGELVMVKEYPEVIRLI 358
Query 67 FDFTMESLRSWERLPNSKHYLLGVWAHVISPLLFYKQKVPNDLELYIERITAAFIMSRM- 125
+FT+ SL+ WE PNS HYLL +W +++ + F K P+ L+ Y IT AFI SR+
Sbjct 359 ANFTITSLQHWEFAPNSVHYLLTLWQRMVASVPFVKSTEPHLLDTYAPEITKAFITSRLD 418
Query 126 CVAEAIADDADN 137
VA + D D+
Sbjct 419 SVAIVVRDHLDD 430
> At5g06120
Length=1059
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 47/135 (34%), Positives = 85/135 (62%), Gaps = 1/135 (0%)
Query 1 FETTAARNKYMSAVILGAFKIIKTRTGLTDDLCYHEFCRLIGKINTSHHLSELCASEPFR 60
F A R+ +++ ++ G +I++T GL D YH FCRL+G+ ++ LSEL E +
Sbjct 280 FTNDATRSNFLAHLMTGTKEILQTGKGLADHDNYHVFCRLLGRFRLNYQLSELVKMEGYG 339
Query 61 EFPQYLFDFTMESLRSWERLPNSKHYLLGVWAHVISPLLFYKQKVPNDLELYIERITAAF 120
E+ Q + +FT++SL+SW+ +S +YLLG+W+ +++ + + K P+ L+ ++ +IT F
Sbjct 340 EWIQLVAEFTLKSLQSWQWASSSVYYLLGMWSRLVASVPYLKGDSPSLLDEFVPKITEGF 399
Query 121 IMSRM-CVAEAIADD 134
I+SR V ++ DD
Sbjct 400 IISRFNSVQASVPDD 414
> Hs11999174
Length=1087
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 78/140 (55%), Gaps = 4/140 (2%)
Query 5 AARNKYMSAVILGAFKIIKTRTGLTDDLCYHEFCRLIGKINTSHHLSELCASEPFREFPQ 64
A R K++S ++ G +I++ L+D YHEFCRL+ ++ +++ L EL E + E +
Sbjct 298 AERAKFLSHLVDGVKRILENPQSLSDPNNYHEFCRLLARLKSNYQLGELVKVENYPEVIR 357
Query 65 YLFDFTMESLRSWERLPNSKHYLLGVWAHVISPLLFYKQKVPNDLELYIERITAAFIMSR 124
+ +FT+ SL+ WE PNS HYLL +W + + + + K P+ LE Y +T A+I SR
Sbjct 358 LIANFTVTSLQHWEFAPNSVHYLLSLWQRLAASVPYVKATEPHMLETYTPEVTKAYITSR 417
Query 125 MCVAEAIADDADNCDWESPL 144
+ I D E PL
Sbjct 418 LESVHIILRDG----LEDPL 433
> CE15680
Length=1078
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 41/138 (29%), Positives = 72/138 (52%), Gaps = 4/138 (2%)
Query 7 RNKYMSAVILGAFKIIKTRTGLTDDLCYHEFCRLIGKINTSHHLSELCASEPFREFPQYL 66
R Y+ ++ G +I L+D +HEFCRLI ++ T++ L EL A + + L
Sbjct 303 RQAYVQKLVEGVVSVIMNPGKLSDQAAFHEFCRLIARLKTNYQLCELIAVPCYSHMLRLL 362
Query 67 FDFTMESLRSWERLPNSKHYLLGVWAHVISPLLFYKQKVPNDLELYIERITAAFIMSRMC 126
+FT++SLR E NS ++L+ W +++ + + + + L +Y I AF+ SR+
Sbjct 363 AEFTVQSLRMMEFSANSTYFLMTFWQRMVTSVPYVRNNDEHLLNVYCPEIMTAFVESRLQ 422
Query 127 VAEAIADDADNCDWESPL 144
E+I + E+PL
Sbjct 423 HVESIVREG----AENPL 436
> 7293158_2
Length=500
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 58/99 (58%), Gaps = 0/99 (0%)
Query 27 GLTDDLCYHEFCRLIGKINTSHHLSELCASEPFREFPQYLFDFTMESLRSWERLPNSKHY 86
GL+D YHEFCRL+ ++ +++ L EL A + E Q + FT+ESL W PNS HY
Sbjct 243 GLSDPDNYHEFCRLLARLKSNYQLGELIAVPCYPEAIQLIAKFTVESLHLWLFAPNSVHY 302
Query 87 LLGVWAHVISPLLFYKQKVPNDLELYIERITAAFIMSRM 125
LL +W +++ + + K P+ L Y + A+I SR+
Sbjct 303 LLTLWQRMVASVPYVKSPDPHLLGTYTPEVIKAYIESRL 341
> Hs22056548_1
Length=583
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 6/54 (11%)
Query 49 HLSEL-CASEPFREFPQYLFDFTMESLR-SWERLPNSK----HYLLGVWAHVIS 96
H+ +L AS P + QYLF FT+E + +W R P ++ L G+W +S
Sbjct 431 HIHQLPAASNPVDQASQYLFAFTLEGQQFTWTRWPRAQGTTAASLRGLWPEKLS 484
> At3g57060
Length=1439
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 1 FETTAARNKYMSAVILGAFKIIKTRTGLTDDLCYHEFCRLIGKINTS-HHLSELCAS 56
F ++ Y+S ++ +F + + T L D CR+IG T H++ + CAS
Sbjct 192 FGSSDLDENYLSFIVKNSFTLFENATILKDAETKDALCRIIGASATKYHYIVQSCAS 248
> At5g55560
Length=313
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query 45 NTSHHLSELCASEPFREFPQYLFDFTMESLRSWERLPNSKHYLLGV-WAHVISPLLFYK 102
NT + ++E+C S RE+ + +M +L+ W SK L G+ + H P + ++
Sbjct 104 NTLNFITEICTSGNLREYRKKHRHVSMRALKKW-----SKQILKGLDYLHTHDPCIIHR 157
> YGL104c
Length=486
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 26/58 (44%), Gaps = 5/58 (8%)
Query 23 KTRTGLTDDLCYHEFCRLIGKINTSHHLSELCASEPFREFPQYLFDFTMESL---RSW 77
+ + +T LCY IG I +HLSEL A + + FD ME R+W
Sbjct 19 QNKHKITKALCYAIIVASIGSIQFGYHLSELNAPQQVLSCSE--FDIPMEGYPYDRTW 74
> YIL049w
Length=253
Score = 28.5 bits (62), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 32/69 (46%), Gaps = 18/69 (26%)
Query 65 YLFDFTMESLRSWERLPNSKHYLLGVWAHVISPLL----FYKQKVPNDLELYIERITAAF 120
Y+ +T S +W HYL+G+W + + L+ YK +PN L + AF
Sbjct 109 YVLHYTSNSRMNWS------HYLVGIWFYSVLLLILNISLYKNSIPNTLNM------NAF 156
Query 121 IMSRMCVAE 129
I+ C+A
Sbjct 157 II--FCIAS 163
> CE05513
Length=378
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 25/53 (47%), Gaps = 7/53 (13%)
Query 84 KHYLL-------GVWAHVISPLLFYKQKVPNDLELYIERITAAFIMSRMCVAE 129
+HYLL GV + +I + +K PN +E +T F+M +AE
Sbjct 131 RHYLLVDDMLKDGVGSILIQNPFYGDRKPPNQFRSSLENVTDLFVMGAALIAE 183
> Hs4757930
Length=398
Score = 27.7 bits (60), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 4/38 (10%)
Query 106 PNDLELYIERITAAFIMSRMCVAEAIADDADNCDWESP 143
P D+ + E + AF + +C E D DN DWE+P
Sbjct 95 PEDVSMKEENLCQAFSDALLCKIE----DIDNEDWENP 128
> At1g29730
Length=940
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query 32 LCYHEFCRLIGKINTSHHLSELCAS--EPFREFPQYLFDFTMESLRSWE 78
L Y+ FC G N H +E+ S EP+ + + +F+ ++ WE
Sbjct 466 LAYYAFCFENGSYNVKLHFAEIQFSDVEPYTKLAKRVFNIYIQGKLIWE 514
Lambda K H
0.325 0.137 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40