bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2668_orf1
Length=138
Score E
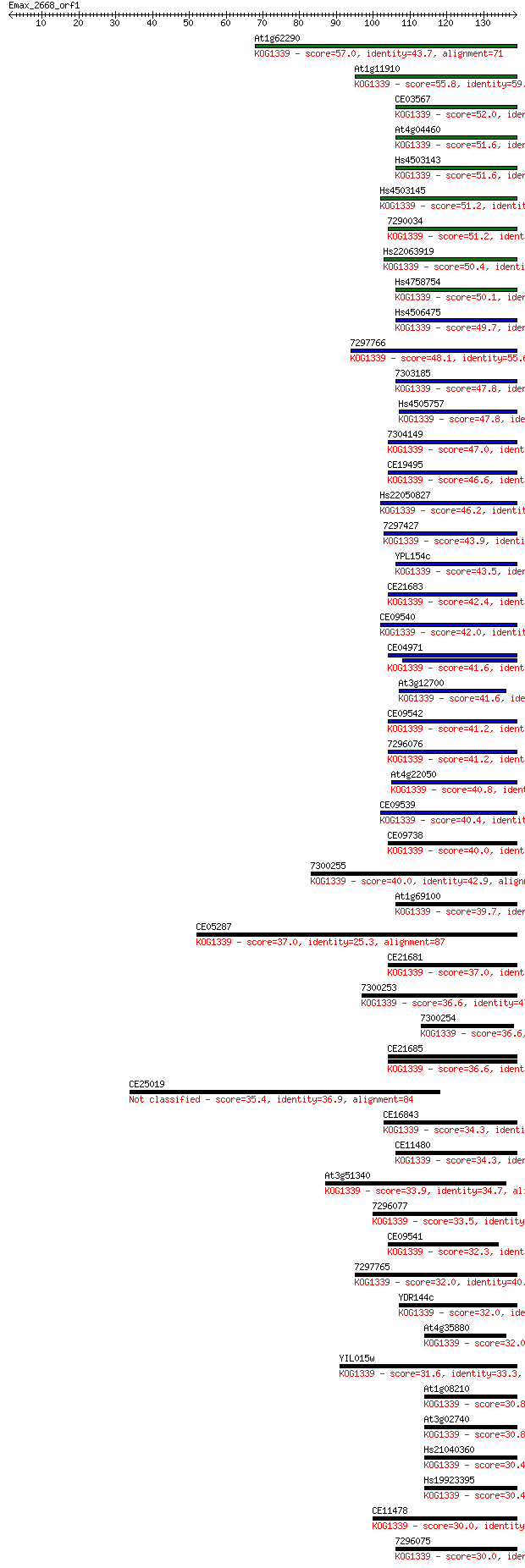
Sequences producing significant alignments: (Bits) Value
At1g62290 57.0 1e-08
At1g11910 55.8 2e-08
CE03567 52.0 3e-07
At4g04460 51.6 4e-07
Hs4503143 51.6 4e-07
Hs4503145 51.2 6e-07
7290034 51.2 6e-07
Hs22063919 50.4 1e-06
Hs4758754 50.1 1e-06
Hs4506475 49.7 2e-06
7297766 48.1 5e-06
7303185 47.8 6e-06
Hs4505757 47.8 6e-06
7304149 47.0 1e-05
CE19495 46.6 2e-05
Hs22050827 46.2 2e-05
7297427 43.9 1e-04
YPL154c 43.5 1e-04
CE21683 42.4 3e-04
CE09540 42.0 4e-04
CE04971 41.6 5e-04
At3g12700 41.6 5e-04
CE09542 41.2 5e-04
7296076 41.2 6e-04
At4g22050 40.8 7e-04
CE09539 40.4 0.001
CE09738 40.0 0.001
7300255 40.0 0.001
At1g69100 39.7 0.002
CE05287 37.0 0.012
CE21681 37.0 0.012
7300253 36.6 0.015
7300254 36.6 0.015
CE21685 36.6 0.016
CE25019 35.4 0.034
CE16843 34.3 0.068
CE11480 34.3 0.074
At3g51340 33.9 0.084
7296077 33.5 0.12
CE09541 32.3 0.24
7297765 32.0 0.35
YDR144c 32.0 0.37
At4g35880 32.0 0.38
YIL015w 31.6 0.43
At1g08210 30.8 0.78
At3g02740 30.8 0.84
Hs21040360 30.4 1.0
Hs19923395 30.4 1.0
CE11478 30.0 1.3
7296075 30.0 1.3
> At1g62290
Length=526
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 42/71 (59%), Gaps = 2/71 (2%)
Query 68 KLDNRYKFTGLGELVSQLIDHHTTMGSVGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRF 127
+L R+ L S L ++ +G G SG L NY ++QY+GEI IGTP ++F
Sbjct 45 RLATRFGSKQEEALRSSLRSYNNNLG--GDSGDADIVPLKNYLDAQYYGEIAIGTPPQKF 102
Query 128 VVVFDTGSSNL 138
V+FDTGSSNL
Sbjct 103 TVIFDTGSSNL 113
> At1g11910
Length=506
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/44 (59%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 95 VGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+G SG L NY ++QY+GEI IGTP ++F VVFDTGSSNL
Sbjct 63 LGDSGDADVVVLKNYLDAQYYGEIAIGTPPQKFTVVFDTGSSNL 106
> CE03567
Length=444
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 22/33 (66%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY ++QYFG I IGTP + F V+FDTGSSNL
Sbjct 86 LRNYMDAQYFGTISIGTPAQNFTVIFDTGSSNL 118
> At4g04460
Length=508
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 21/33 (63%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY ++QY+G+I IGTP ++F V+FDTGSSNL
Sbjct 79 LKNYLDAQYYGDITIGTPPQKFTVIFDTGSSNL 111
> Hs4503143
Length=412
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY ++QY+GEI IGTP + F VVFDTGSSNL
Sbjct 71 LKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNL 103
> Hs4503145
Length=396
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 21/37 (56%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 102 ARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
A++ L+NY + +YFG I IG+P + F V+FDTGSSNL
Sbjct 66 AKEPLINYLDMEYFGTISIGSPPQNFTVIFDTGSSNL 102
> 7290034
Length=407
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ L NY N QY+G I IGTPG+ F+V FDTGSSNL
Sbjct 67 EHLSNYDNFQYYGNISIGTPGQDFLVQFDTGSSNL 101
> Hs22063919
Length=267
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 103 RQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q L NY + +YFG I IGTP + F VVFDTGSSNL
Sbjct 65 EQPLENYLDMEYFGTIGIGTPAQDFTVVFDTGSSNL 100
> Hs4758754
Length=420
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/33 (66%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY + QYFGEI +GTP + F V FDTGSSNL
Sbjct 70 LSNYRDVQYFGEIGLGTPPQNFTVAFDTGSSNL 102
> Hs4506475
Length=406
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/33 (66%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY ++QY+GEI IGTP + F VVFDTGSSN+
Sbjct 78 LTNYMDTQYYGEIGIGTPPQTFKVVFDTGSSNV 110
> 7297766
Length=391
Score = 48.1 bits (113), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/45 (55%), Positives = 33/45 (73%), Gaps = 2/45 (4%)
Query 94 SVGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
SV SSG A + L N N++Y+G I IGTP +RF ++FDTGS+NL
Sbjct 58 SVSSSG--ATENLHNSMNNEYYGVIAIGTPEQRFNILFDTGSANL 100
> 7303185
Length=404
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 22/33 (66%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY ++QYFG I IGTP + F V+FDTGSSNL
Sbjct 77 LSNYLDAQYFGPITIGTPPQTFKVIFDTGSSNL 109
> Hs4505757
Length=388
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 107 LNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ Y ++ YFGEI IGTP + F+V+FDTGSSNL
Sbjct 66 MAYMDAAYFGEISIGTPPQNFLVLFDTGSSNL 97
> 7304149
Length=392
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ L NY ++QY+G I IG+P + F VVFDTGSSNL
Sbjct 63 EPLSNYMDAQYYGPIAIGSPPQNFRVVFDTGSSNL 97
> CE19495
Length=398
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ L +Y N+QY+G + IGTP + F V+FDTGSSNL
Sbjct 59 EGLSDYSNAQYYGPVTIGTPPQNFQVLFDTGSSNL 93
> Hs22050827
Length=160
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/37 (56%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 102 ARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
A L + ++QYFGEI +GTP + F V FDTGSSNL
Sbjct 66 ASVPLSKFLDAQYFGEIGLGTPPQNFTVAFDTGSSNL 102
> 7297427
Length=372
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 103 RQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
++L N N Y+G I IGTP + F V+FD+GSSNL
Sbjct 58 EEQLSNSMNMAYYGAISIGTPAQSFKVLFDSGSSNL 93
> YPL154c
Length=405
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/33 (57%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NY N+QY+ +I +GTP + F V+ DTGSSNL
Sbjct 83 LTNYLNAQYYTDITLGTPPQNFKVILDTGSSNL 115
> CE21683
Length=320
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q ++Y + Y G I +GTPG+ +V DTGSSNL
Sbjct 59 QPFIDYMDDFYLGNISVGTPGQTLTLVLDTGSSNL 93
> CE09540
Length=395
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 102 ARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
A Q + ++ + +Y G I IGTP ++F+VV DTGS+NL
Sbjct 61 APQNVNDFGDVEYLGNITIGTPPQQFIVVLDTGSANL 97
> CE04971
Length=709
Score = 41.6 bits (96), Expect = 5e-04, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q + +Y + Y G I IGTP ++F V+ DTGSSNL
Sbjct 344 QYVNDYEDEAYVGNITIGTPQQQFKVILDTGSSNL 378
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 108 NYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+Y ++ Y G+I +G+P + F V+ DTGSSN
Sbjct 34 DYEHAGYVGKITVGSPPQEFRVIMDTGSSNF 64
> At3g12700
Length=439
Score = 41.6 bits (96), Expect = 5e-04, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 107 LNYHNSQYFGEIKIGTPGRRFVVVFDTGS 135
++Y +QYF EI++GTP ++F VV DTGS
Sbjct 77 IDYGTAQYFTEIRVGTPAKKFRVVVDTGS 105
> CE09542
Length=389
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q + ++ + +Y G I IGTP + F+VV DTGSSNL
Sbjct 61 QNVNDFGDFEYLGNITIGTPDQGFIVVLDTGSSNL 95
> 7296076
Length=405
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
++L N N Y+G I IGTP + F VVFDTGS+NL
Sbjct 83 EELGNSMNMYYYGLIGIGTPEQYFKVVFDTGSANL 117
> At4g22050
Length=336
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 105 KLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+L N + Y+G+I+IG PG+ F V+FDTGSS+L
Sbjct 37 QLKNVKDFLYYGKIQIGNPGQTFTVLFDTGSSSL 70
> CE09539
Length=393
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 102 ARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
A Q + ++ + +Y G I IGTP + F+VV DTGSSNL
Sbjct 61 APQNVNDFGDFEYLGNITIGTPPQPFLVVLDTGSSNL 97
> CE09738
Length=389
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q +Y +Y G I +GTP + F+VV DTGS+NL
Sbjct 62 QSTTDYVYYEYMGNITVGTPDQNFIVVLDTGSANL 96
> 7300255
Length=395
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 83 SQLIDHHTTMGS--VGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
S L+ + +G V S A + L N N +Y G I IG+PG+ F ++FDTGS+NL
Sbjct 48 SSLLAKYNVVGGQEVTSRNGGATETLDNRLNLEYAGPISIGSPGQPFNMLFDTGSANL 105
> At1g69100
Length=343
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L N+ ++GEI +G+P ++F VVFDTGS++L
Sbjct 39 LKNFDGVVFYGEISVGSPPQKFNVVFDTGSTDL 71
> CE05287
Length=428
Score = 37.0 bits (84), Expect = 0.012, Method: Composition-based stats.
Identities = 22/88 (25%), Positives = 46/88 (52%), Gaps = 7/88 (7%)
Query 52 TADLHT-NLLREPPMTIKLDNRYKFTGLGELVSQLIDHHTTMGSVGSSGTMARQKLLNYH 110
+A++H N+ P M +++ + K + ++L+ + + SS +++Y
Sbjct 15 SAEVHQFNIGYRPNMRQRMNAKGKLAEYEKERNELLSKKSLQLASSSS------PVIDYE 68
Query 111 NSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ Y +I +G+P + FV+ D+GSSNL
Sbjct 69 DMAYMVQISLGSPAQNFVLFIDSGSSNL 96
> CE21681
Length=396
Score = 37.0 bits (84), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q ++Y + Y G I +GTP + VV DTGSSNL
Sbjct 58 QPFVDYFDDFYLGNITLGTPPQPATVVLDTGSSNL 92
> 7300253
Length=465
Score = 36.6 bits (83), Expect = 0.015, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 27/42 (64%), Gaps = 3/42 (7%)
Query 97 SSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
SSGT L N N +Y ++ IGTP ++F V+ DTGSSN+
Sbjct 136 SSGTAT---LKNTANMEYTCKMNIGTPKQKFTVLPDTGSSNI 174
> 7300254
Length=309
Score = 36.6 bits (83), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 113 QYFGEIKIGTPGRRFVVVFDTGSSN 137
+Y+G I +G P + F V+FDTGSSN
Sbjct 2 EYYGTIAMGNPRQNFTVIFDTGSSN 26
> CE21685
Length=829
Score = 36.6 bits (83), Expect = 0.016, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q L++Y++ Y I +GTP + VV DT S+NL
Sbjct 58 QPLIDYYDDMYLANITVGTPPQPASVVLDTASANL 92
Score = 33.5 bits (75), Expect = 0.14, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
Q + +Y + Y G +GTP + +V DTGS+N+
Sbjct 493 QPISDYSDEVYLGNFTVGTPPQPVSLVLDTGSANM 527
> CE25019
Length=391
Score = 35.4 bits (80), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 45/96 (46%), Gaps = 21/96 (21%)
Query 34 RHRFLSETLEEPEDVML---KTADLHTNLLREPPM----TIKLDNRYKFT-----GLGEL 81
R +FLSE LE PE++ML K ADL + PP+ T++L FT G+G++
Sbjct 28 RQKFLSENLENPEEIMLNGIKLADLKIGPVDMPPLDHRQTVQLS---MFTVKTRRGIGQI 84
Query 82 VSQLIDHHTTMGSVGSSGTMARQKLLNYHNSQYFGE 117
+ +L G S + R +LN GE
Sbjct 85 LFEL------AALFGISRKLGRVPVLNRLQEPLIGE 114
> CE16843
Length=394
Score = 34.3 bits (77), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 5/41 (12%)
Query 103 RQKLLNYHNSQYFGEIKIGTP-----GRRFVVVFDTGSSNL 138
Q + ++ + YFG I +GTP + F+VV DTGSSN+
Sbjct 55 HQHVADFRDFAYFGNITLGTPIESTAEQTFLVVLDTGSSNV 95
> CE11480
Length=474
Score = 34.3 bits (77), Expect = 0.074, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
++ + Y ++IGTP + F V FDT SSNL
Sbjct 145 FFDHFDEYYTAGVRIGTPAQHFQVAFDTTSSNL 177
> At3g51340
Length=518
Score = 33.9 bits (76), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 87 DHHTTMGSVGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGS 135
+ T + S+GS+ T+A LN+ ++ + +GTP F+V DTGS
Sbjct 68 NEETPLTSIGSNLTLA----LNFLGFLHYANVSLGTPATWFLVALDTGS 112
> 7296077
Length=418
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 1/40 (2%)
Query 100 TMARQKLLNYHNSQYFGEIKIGTPGRRFVVVF-DTGSSNL 138
T++++ L+N HN++Y+ GTP + V + DT S+NL
Sbjct 78 TVSKENLINSHNTEYYVTAGFGTPKSQPVTLLVDTASANL 117
> CE09541
Length=350
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 104 QKLLNYHNSQYFGEIKIGTPGRRFVVVFDT 133
Q + ++ + +Y G I IGTP + F+VV DT
Sbjct 62 QNVNDFADFEYLGNITIGTPDQSFIVVLDT 91
> 7297765
Length=423
Score = 32.0 bits (71), Expect = 0.35, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 95 VGSSGTMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
V + Q L N N +Y + IGTP + F + FDTGSS+L
Sbjct 55 VDRQSSQTTQVLANGFNLEYTIRLCIGTPPQCFNLQFDTGSSDL 98
> YDR144c
Length=596
Score = 32.0 bits (71), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 107 LNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L NS Y E+ IGTP ++ V+ DTGSS+L
Sbjct 74 LTNQNSFYSVELDIGTPPQKVTVLVDTGSSDL 105
> At4g35880
Length=455
Score = 32.0 bits (71), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 114 YFGEIKIGTPGRRFVVVFDTGS 135
++ +K+GTPG RF+V DTGS
Sbjct 107 HYTTVKLGTPGMRFMVALDTGS 128
> YIL015w
Length=587
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 91 TMGSVGSSGTMARQKLLNYHNSQYFGE-IKIGTPGRRFVVVFDTGSSNL 138
T+ ++ + GT + LL + Y+ + IGTP + V+FDTGS++
Sbjct 21 TITALTNDGTGHLEFLLQHEEEMYYATTLDIGTPSQSLTVLFDTGSADF 69
> At1g08210
Length=566
Score = 30.8 bits (68), Expect = 0.78, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 114 YFGEIKIGTPGRRFVVVFDTGSSNL 138
Y+ ++K+GTP R F V DTGS L
Sbjct 132 YYTKVKLGTPPREFNVQIDTGSDVL 156
> At3g02740
Length=488
Score = 30.8 bits (68), Expect = 0.84, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 114 YFGEIKIGTPGRRFVVVFDTGSSNL 138
YF +I +GTP R F V DTGS L
Sbjct 85 YFAKIGLGTPSRDFHVQVDTGSDIL 109
> Hs21040360
Length=468
Score = 30.4 bits (67), Expect = 1.0, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 114 YFGEIKIGTPGRRFVVVFDTGSSNL 138
Y+ E+ IGTP ++ ++ DTGSSN
Sbjct 92 YYLEMLIGTPPQKLQILVDTGSSNF 116
> Hs19923395
Length=518
Score = 30.4 bits (67), Expect = 1.0, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 114 YFGEIKIGTPGRRFVVVFDTGSSNL 138
Y+ E+ IGTP ++ ++ DTGSSN
Sbjct 92 YYLEMLIGTPPQKLQILVDTGSSNF 116
> CE11478
Length=383
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 100 TMARQKLLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
+ + + ++ + Y ++IGTP + F V DT SSNL
Sbjct 48 STGNESIYDHFDEYYTVSVRIGTPAQHFEVALDTTSSNL 86
> 7296075
Length=410
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 106 LLNYHNSQYFGEIKIGTPGRRFVVVFDTGSSNL 138
L N +N++Y+ + G P + V+ DTGS+NL
Sbjct 82 LENLYNTEYYTTLGFGNPPQDLKVLIDTGSANL 114
Lambda K H
0.317 0.133 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40