bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2645_orf1
Length=104
Score E
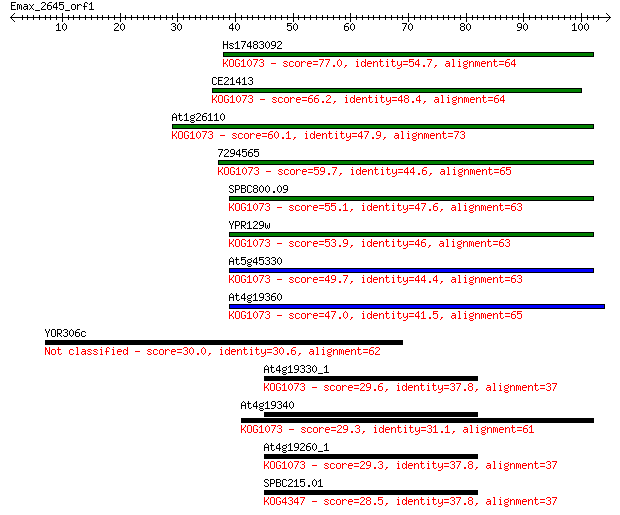
Sequences producing significant alignments: (Bits) Value
Hs17483092 77.0 8e-15
CE21413 66.2 1e-11
At1g26110 60.1 1e-09
7294565 59.7 1e-09
SPBC800.09 55.1 3e-08
YPR129w 53.9 8e-08
At5g45330 49.7 1e-06
At4g19360 47.0 9e-06
YOR306c 30.0 0.98
At4g19330_1 29.6 1.2
At4g19340 29.3 1.7
At4g19260_1 29.3 1.9
SPBC215.01 28.5 3.5
> Hs17483092
Length=463
Score = 77.0 bits (188), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 35/64 (54%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 38 PYTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVPIPVPPSPEIYNSV 97
PY GS+ISLI+ ++IR+EGIL +I+T STV+L V+SFGTEDRP P+PP E++ +
Sbjct 6 PYIGSKISLISKAEIRYEGILYTIDTENSTVALAKVRSFGTEDRPTDRPIPPRDEVFEYI 65
Query 98 VFYG 101
+F G
Sbjct 66 IFRG 69
> CE21413
Length=340
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 31/64 (48%), Positives = 45/64 (70%), Gaps = 0/64 (0%)
Query 36 ELPYTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVPIPVPPSPEIYN 95
+ PY GS+ISLI+ DIR+EGIL +++T ST++L V+SFGTE RP PV ++Y
Sbjct 4 QTPYIGSKISLISKLDIRYEGILYTVDTNDSTIALAKVRSFGTEKRPTANPVAARDDVYE 63
Query 96 SVVF 99
++F
Sbjct 64 YIIF 67
> At1g26110
Length=643
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 29 GSQGAGEELPYTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVPIP-V 87
GS+ + Y GS ISL + S+IR+EGIL +INT S++ L+ V+SFGTE R P V
Sbjct 7 GSKSSSAADSYVGSLISLTSKSEIRYEGILYNINTDESSIGLQNVRSFGTEGRKKDGPQV 66
Query 88 PPSPEIYNSVVFYG 101
PPS ++Y ++F G
Sbjct 67 PPSDKVYEYILFRG 80
> 7294565
Length=652
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 37 LPYTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVPIPVPPSPEIYNS 96
LP GS+ISLI+ +DIR+EG L +++ T++L V+SFGTEDR + P +IY+
Sbjct 5 LPELGSKISLISKADIRYEGRLYTVDPQECTIALSSVRSFGTEDRDTQFQIAPQSQIYDY 64
Query 97 VVFYG 101
++F G
Sbjct 65 ILFRG 69
> SPBC800.09
Length=426
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 43/65 (66%), Gaps = 2/65 (3%)
Query 39 YTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVP--IPVPPSPEIYNS 96
+ GSRISLI+ SDIR+ GIL IN+ ST++L+ V+ GTE R +PPS +++
Sbjct 4 FIGSRISLISKSDIRYVGILQDINSQDSTLALKHVRWCGTEGRKQDPSQEIPPSDNVFDY 63
Query 97 VVFYG 101
+VF G
Sbjct 64 IVFRG 68
> YPR129w
Length=349
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 42/65 (64%), Gaps = 2/65 (3%)
Query 39 YTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPV--PIPVPPSPEIYNS 96
Y G ISLI+ +D R+ G+L+ I++ TV+L+ V+ FGTE R P + P+P +YNS
Sbjct 4 YIGKTISLISVTDNRYVGLLEDIDSEKGTVTLKEVRCFGTEGRKNWGPEEIYPNPTVYNS 63
Query 97 VVFYG 101
V F G
Sbjct 64 VKFNG 68
> At5g45330
Length=571
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Query 39 YTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVPIP-VPPSPEIYNSV 97
+ GS ISLI+ +IR+EGIL +N ST+ L+ V+S GTE R P +PP ++Y+ +
Sbjct 33 FIGSFISLISKYEIRYEGILYHLNVQDSTLGLKNVRSCGTEGRKKDGPQIPPCDKVYDYI 92
Query 98 VFYG 101
+F G
Sbjct 93 LFRG 96
> At4g19360
Length=268
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 39 YTGSRISLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDRPVP-IPVPPSPEIYNSV 97
Y GS ++LI DIR+EGIL +N ST+ L+ V +GTE R + +PP +I N +
Sbjct 27 YIGSFVTLIANFDIRYEGILCFLNLQESTLGLQNVVCYGTEGRNQNGVQIPPDTKIQNYI 86
Query 98 VFYGKT 103
+F G
Sbjct 87 LFNGNN 92
> YOR306c
Length=521
Score = 30.0 bits (66), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 27/62 (43%), Gaps = 1/62 (1%)
Query 7 CCSLNRPAEPLSSQRGEEVFTMGSQGAGEELPYTGSRISLITTSDIRHEGILDSINTVAS 66
CC E + F +G G +P TG+ IS+ TT+D +H I + T S
Sbjct 444 CCGQISKTEEFGKRYSTMYFVVGF-GTLVGIPITGAIISIKTTADYQHYIIFSGLATFVS 502
Query 67 TV 68
V
Sbjct 503 AV 504
> At4g19330_1
Length=167
Score = 29.6 bits (65), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 45 SLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDR 81
SLITT D+R EG++ + S + ++ +GTE R
Sbjct 130 SLITTEDVRIEGVISHVKFHDSMIFMKNCMCYGTEGR 166
> At4g19340
Length=232
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 45 SLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDR 81
SLITT D+R EG++ + S + ++ +GTE R
Sbjct 134 SLITTEDVRIEGVISHVKFHDSMIFMKNCMCYGTEGR 170
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 41 GSRISLITTSDIRHEGILDSINTVASTVSLR-FVQSFG--TEDRPVPIPVPPSPEIYNSV 97
G +++++ +DIR+EG++ +N S + L+ V+ +G E+ E+++ +
Sbjct 21 GKFVAVLSNNDIRYEGVISLLNLQDSKLGLQNVVRVYGREVENDNEQRVFQVLKEVHSHM 80
Query 98 VFYG 101
VF G
Sbjct 81 VFRG 84
> At4g19260_1
Length=233
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 45 SLITTSDIRHEGILDSINTVASTVSLRFVQSFGTEDR 81
SLITT D+R EG++ + S + ++ +GTE R
Sbjct 134 SLITTEDVRIEGVISHVKFHDSMIFMKNCMCYGTEGR 170
> SPBC215.01
Length=827
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 45 SLITTSDIRH-EGILDSINTVASTVSLRFVQSFGTEDR 81
SLI +H EG+++SI + A +R +Q++GT R
Sbjct 483 SLIEDERKKHYEGVMNSIESFAKRTQIRSLQNYGTLTR 520
Lambda K H
0.315 0.133 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40