bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
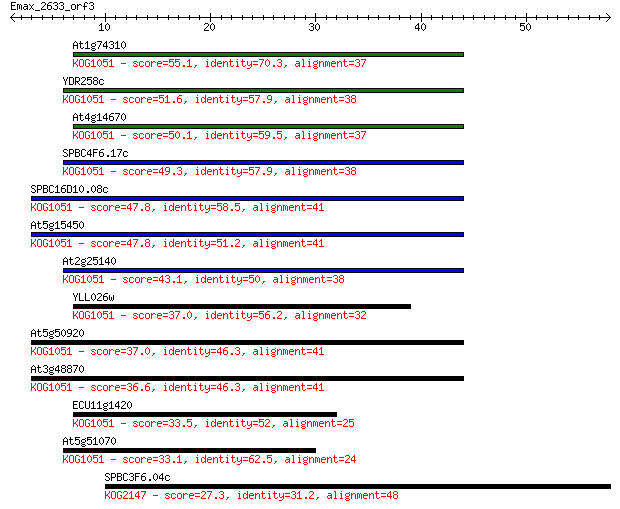
Query= Emax_2633_orf3
Length=57
Score E
Sequences producing significant alignments: (Bits) Value
At1g74310 55.1 3e-08
YDR258c 51.6 4e-07
At4g14670 50.1 9e-07
SPBC4F6.17c 49.3 2e-06
SPBC16D10.08c 47.8 5e-06
At5g15450 47.8 6e-06
At2g25140 43.1 1e-04
YLL026w 37.0 0.008
At5g50920 37.0 0.008
At3g48870 36.6 0.012
ECU11g1420 33.5 0.096
At5g51070 33.1 0.12
SPBC3F6.04c 27.3 6.6
> At1g74310
Length=911
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 26/37 (70%), Positives = 30/37 (81%), Gaps = 0/37 (0%)
Query 7 LPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
LPDKAIDL+DEACA RVQ+DS+PE +D LER QL
Sbjct 387 LPDKAIDLVDEACANVRVQLDSQPEEIDNLERKRMQL 423
> YDR258c
Length=811
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/38 (57%), Positives = 32/38 (84%), Gaps = 0/38 (0%)
Query 6 FLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
FLPDKAIDL+DEACA+ R+Q +SKP+ + +L+R I ++
Sbjct 320 FLPDKAIDLVDEACAVLRLQHESKPDEIQKLDRAIMKI 357
> At4g14670
Length=623
Score = 50.1 bits (118), Expect = 9e-07, Method: Composition-based stats.
Identities = 22/37 (59%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 7 LPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
LPDKAIDL+DE+CA + Q+D +PE +D LER + QL
Sbjct 352 LPDKAIDLVDESCAHVKAQLDIQPEEIDSLERKVMQL 388
> SPBC4F6.17c
Length=803
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/38 (57%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 6 FLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
FLPDKAIDL+DEAC+ R+Q +SKP+ + RL+R I +
Sbjct 318 FLPDKAIDLVDEACSSLRLQQESKPDELRRLDRQIMTI 355
> SPBC16D10.08c
Length=905
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 3 SSLFLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
+S LPD AIDL+DEA A RV +S+PEV+D LER + QL
Sbjct 386 TSRRLPDSAIDLVDEAAAAVRVTRESQPEVLDNLERKLRQL 426
> At5g15450
Length=968
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 21/41 (51%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 3 SSLFLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
S FLPDKAIDL+DEA A ++++ SKP +D L+R + +L
Sbjct 458 SGRFLPDKAIDLVDEAAAKLKMEITSKPTALDELDRSVIKL 498
> At2g25140
Length=874
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 6 FLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
FLPDKAIDL+DEA A ++++ SKP +D ++R + +L
Sbjct 376 FLPDKAIDLVDEAGAKLKMEITSKPTELDGIDRAVIKL 413
> YLL026w
Length=908
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 7 LPDKAIDLMDEACAIARVQVDSKPEVMDRLER 38
LPD A+DL+D +CA V DSKPE +D ER
Sbjct 388 LPDSALDLVDISCAGVAVARDSKPEELDSKER 419
> At5g50920
Length=929
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 3 SSLFLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
S FLPDKAIDL+DEA + R++ PE LE+ + Q+
Sbjct 477 SDRFLPDKAIDLIDEAGSRVRLRHAQVPEEARELEKELRQI 517
> At3g48870
Length=952
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 3 SSLFLPDKAIDLMDEACAIARVQVDSKPEVMDRLERFIFQL 43
S FLPDKAIDL+DEA + R++ PE LE+ + Q+
Sbjct 498 SDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELEKQLRQI 538
> ECU11g1420
Length=851
Score = 33.5 bits (75), Expect = 0.096, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 7 LPDKAIDLMDEACAIARVQVDSKPE 31
LPD AIDL+D ACA + ++S+P+
Sbjct 373 LPDVAIDLLDTACASVMIALESEPQ 397
> At5g51070
Length=945
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 15/24 (62%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 6 FLPDKAIDLMDEACAIARVQVDSK 29
FLPDKAIDL+DEA + AR++ K
Sbjct 499 FLPDKAIDLIDEAGSRARIEAFRK 522
> SPBC3F6.04c
Length=827
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 4/52 (7%)
Query 10 KAIDLMDEACAIARVQ----VDSKPEVMDRLERFIFQLAFFVLHISPMPLIT 57
K +D D ++R++ V P+ RLE F L +LH++ P+I+
Sbjct 424 KGLDYKDYPTVVSRIRTLHHVKLHPDNKSRLENFSVILLQHILHLTRQPMIS 475
Lambda K H
0.330 0.141 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191047922
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40