bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2612_orf2
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
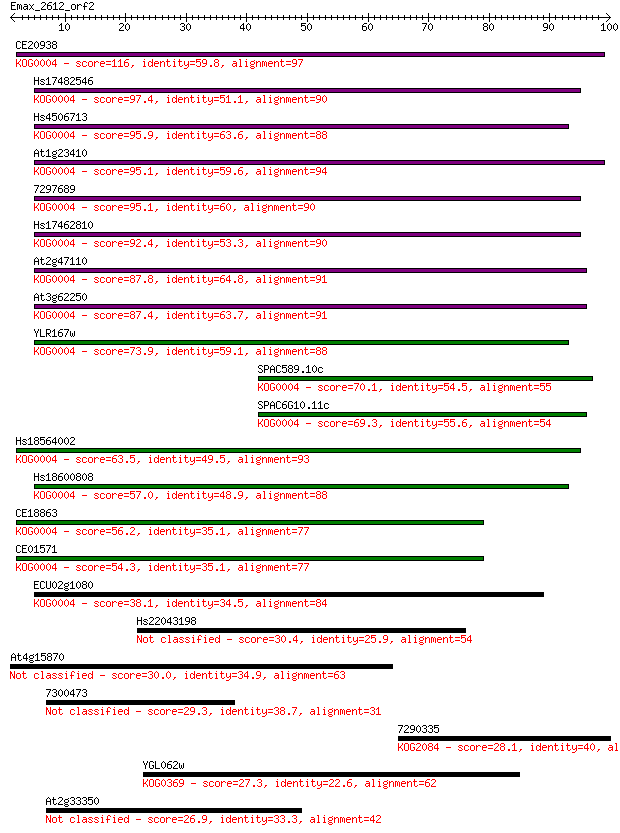
CE20938 116 1e-26
Hs17482546 97.4 5e-21
Hs4506713 95.9 1e-20
At1g23410 95.1 3e-20
7297689 95.1 3e-20
Hs17462810 92.4 2e-19
At2g47110 87.8 4e-18
At3g62250 87.4 5e-18
YLR167w 73.9 6e-14
SPAC589.10c 70.1 9e-13
SPAC6G10.11c 69.3 2e-12
Hs18564002 63.5 8e-11
Hs18600808 57.0 8e-09
CE18863 56.2 1e-08
CE01571 54.3 5e-08
ECU02g1080 38.1 0.004
Hs22043198 30.4 0.77
At4g15870 30.0 1.2
7300473 29.3 1.7
7290335 28.1 4.6
YGL062w 27.3 6.2
At2g33350 26.9 9.0
> CE20938
Length=163
Score = 116 bits (290), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 58/97 (59%), Positives = 74/97 (76%), Gaps = 0/97 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ E+T++ +LELLG KKRK+K YT PKK K K KKVKLAVLK+YK+DEN +TRL
Sbjct 52 DCGIDSEATIYVNLELLGGAKKRKKKVYTTPKKNKRKPKKVKLAVLKYYKIDENGKITRL 111
Query 62 RKECPSKTCGSGVFMAQHENRNYCGRCGITYILQSGN 98
RKEC +CG GVFMAQH NR+YCGRC T ++ +
Sbjct 112 RKECQQPSCGGGVFMAQHANRHYCGRCHDTLVVDTAT 148
> Hs17482546
Length=129
Score = 97.4 bits (241), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 64/90 (71%), Gaps = 1/90 (1%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++ES + L + G K+RK+++ T K KHK KKVKLAVLK+YKVDEN ++ LR+
Sbjct 35 IQKESPLHLVLRIRGGAKERKKRSLT-CKTNKHKRKKVKLAVLKYYKVDENGKISHLRRG 93
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYIL 94
CPS CG+ VFMA H +R+YCG+C +TY
Sbjct 94 CPSDECGAAVFMASHFDRHYCGKCCLTYCF 123
> Hs4506713
Length=156
Score = 95.9 bits (237), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 71/88 (80%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN ++RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKISRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CPS CG+GVFMA H +R+YCG+C +TY
Sbjct 121 CPSDECGAGVFMASHFDRHYCGKCCLTY 148
> At1g23410
Length=156
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 56/94 (59%), Positives = 72/94 (76%), Gaps = 0/94 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KH +KKVKLAVL+FYKVD + V RL+KE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHTHKKVKLAVLQFYKVDGSGKVQRLKKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQSGN 98
CPS +CG G FMA H +R+YCG+CG TY+ + +
Sbjct 121 CPSVSCGPGTFMASHFDRHYCGKCGTTYVFKKAD 154
> 7297689
Length=156
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K Y+ PKK KHK KKVKLAVLK+YKVDEN + RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKNYSTPKKIKHKRKKVKLAVLKYYKVDENGKIHRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYIL 94
CP + CG+GVFMA HE+R+YCG+C +T++
Sbjct 121 CPGENCGAGVFMAAHEDRHYCGKCNLTFVF 150
> Hs17462810
Length=130
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/90 (53%), Positives = 64/90 (71%), Gaps = 3/90 (3%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK K+YT PK KHK KK+KLAVLK+YKV+EN + L +E
Sbjct 38 IQKESTLHLGLRLCGGAKKRK-KSYTTPKNNKHKRKKMKLAVLKYYKVEENGKIRHLCRE 96
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYIL 94
C S G+GVFMA + +R+YCG+C +TY
Sbjct 97 CHSD--GAGVFMASYFDRHYCGKCCLTYCF 124
> At2g47110
Length=157
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 59/91 (64%), Positives = 74/91 (81%), Gaps = 0/91 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KHK+KKVKLAVL+FYKVD + V RLRKE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHKHKKVKLAVLQFYKVDGSGKVQRLRKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQ 95
CP+ TCG+G FMA H +R+YCG+CG+TY+ Q
Sbjct 121 CPNATCGAGTFMASHFDRHYCGKCGLTYVYQ 151
> At3g62250
Length=157
Score = 87.4 bits (215), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 58/91 (63%), Positives = 74/91 (81%), Gaps = 0/91 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KHK+KKVKLAVL+FYK+D + V RLRKE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHKHKKVKLAVLQFYKIDGSGKVQRLRKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQ 95
CP+ TCG+G FMA H +R+YCG+CG+TY+ Q
Sbjct 121 CPNATCGAGTFMASHFDRHYCGKCGLTYVYQ 151
> YLR167w
Length=152
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 52/88 (59%), Positives = 67/88 (76%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G GKKRK+K YT PKK KHK+KKVKLAVL +YKVD VT+LR+E
Sbjct 61 IQKESTLHLVLRLRGGGKKRKKKVYTTPKKIKHKHKKVKLAVLSYYKVDAEGKVTKLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
C + TCG+GVF+A H++R YCG+C Y
Sbjct 121 CSNPTCGAGVFLANHKDRLYCGKCHSVY 148
> SPAC589.10c
Length=150
Score = 70.1 bits (170), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 30/55 (54%), Positives = 44/55 (80%), Gaps = 2/55 (3%)
Query 42 VKLAVLKFYKVDENNNVTRLRKECPSKTCGSGVFMAQHENRNYCGRCGITYILQS 96
V+LAVLK+YKV+++ +V RLR+ECP+ CG+ FMA H++R YCGRC +T L++
Sbjct 98 VELAVLKYYKVEDDGSVKRLRRECPN--CGASTFMANHKDRLYCGRCHLTLKLEA 150
> SPAC6G10.11c
Length=150
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/54 (55%), Positives = 43/54 (79%), Gaps = 2/54 (3%)
Query 42 VKLAVLKFYKVDENNNVTRLRKECPSKTCGSGVFMAQHENRNYCGRCGITYILQ 95
V+LAVLK+YKV+++ +V RLR+ECP+ CG+ FMA H++R YCGRC +T L+
Sbjct 98 VELAVLKYYKVEDDGSVKRLRRECPN--CGASTFMANHKDRLYCGRCHLTLKLE 149
> Hs18564002
Length=141
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 46/93 (49%), Positives = 57/93 (61%), Gaps = 19/93 (20%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ +++EST+ L L G KKRK K+YT PKK KHK KKVKLAVLK+YKVDEN
Sbjct 58 DYSIQKESTLHLVLRLRGGDKKRK-KSYTTPKKNKHKRKKVKLAVLKYYKVDENG----- 111
Query 62 RKECPSKTCGSGVFMAQHENRNYCGRCGITYIL 94
FMA H +R+YCG+C +TY L
Sbjct 112 -------------FMASHFDRHYCGKCCLTYCL 131
> Hs18600808
Length=138
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 43/88 (48%), Positives = 56/88 (63%), Gaps = 18/88 (20%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT P+K KHK KKVKLA+LK+YKVDEN
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPRKNKHKRKKVKLALLKYYKVDENG-------- 112
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
FMA H +R+YCG+C +TY
Sbjct 113 ----------FMASHFDRHYCGKCCLTY 130
> CE18863
Length=114
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ +T++ + ELLG KKRK+K Y PKK +K K++K+DEN ++RL
Sbjct 25 DCGIKAAATIYVNCELLGGAKKRKKKVYIIPKKNNISQRKSSSPSPKYFKIDENGKISRL 84
Query 62 RKECPSKTCGSGVFMAQ 78
P+ + FM Q
Sbjct 85 PTGIPTVSIQREAFMTQ 101
> CE01571
Length=142
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ +T++ + ELLG KKRK+K YT PKK +K K++K+DEN ++RL
Sbjct 53 DCGIKAAATIYVNCELLGGAKKRKKKVYTIPKKNNISQRKSSSPSPKYFKIDENGKISRL 112
Query 62 RKECPSKTCGSGVFMAQ 78
+ + FM Q
Sbjct 113 PTGILTASIQREAFMTQ 129
> ECU02g1080
Length=152
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 11/88 (12%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDEN----NNVTR 60
+++EST+ L L G G K+K+K ++ PKK K KK L+ L + V ++ N V+
Sbjct 61 IQKESTLHLVLRLRGGGAKKKKKDFSTPKKVKEPRKKRGLSWLDNFTVTDDKVKMNGVS- 119
Query 61 LRKECPSKTCGSGVFMAQHENRNYCGRC 88
+ CG+ VF + ++ CGRC
Sbjct 120 ------CEVCGTEVFCGKIQSGFKCGRC 141
> Hs22043198
Length=275
Score = 30.4 bits (67), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 21/54 (38%), Gaps = 0/54 (0%)
Query 22 KKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKECPSKTCGSGVF 75
+KR R +P+ +H + + KF + RLR P CG G
Sbjct 111 QKRTRVQLRRPRASRHTRTGICMGAQKFQDLTLERPARRLRLPAPHDLCGLGAL 164
> At4g15870
Length=583
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 10/73 (13%)
Query 1 EEAGVEEESTVFQSLELLGAGKKRKRKTY----TKPKKQKHKNKKVKLAV------LKFY 50
E GVE V + LG G+ + + Y ++PK + + K+ + V +K Y
Sbjct 445 EIGGVEVTMYVSIACSFLGLGQSSREQAYKWLKSRPKFVEAQAKRARFDVNAIMYYMKQY 504
Query 51 KVDENNNVTRLRK 63
KV E TRL+K
Sbjct 505 KVTEEETFTRLQK 517
> 7300473
Length=1426
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 7 EESTVFQSLELLGAGKKRKRKTYTKPKKQKH 37
E T +Q L L+G+ K+ +K TKP++ H
Sbjct 706 EAPTPYQQLPLIGSNKRPSKKPVTKPEQPSH 736
> 7290335
Length=430
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 6/37 (16%)
Query 65 CPSKTCGSGVFMAQHENRNYCGRC--GITYILQSGNN 99
CP++ CG+G+ + +RN C RC GI+ L++ N
Sbjct 296 CPNRNCGAGISV----DRNNCPRCDAGISPKLRNAFN 328
> YGL062w
Length=1178
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 14/62 (22%), Positives = 30/62 (48%), Gaps = 8/62 (12%)
Query 23 KRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKECPSKTCGSGVFMAQHENR 82
+R+ + + K ++V+ A+L + +L KEC + G+ F+ ++NR
Sbjct 252 QRRHQKVVEVAPAKTLPREVRDAIL--------TDAVKLAKECGYRNAGTAEFLVDNQNR 303
Query 83 NY 84
+Y
Sbjct 304 HY 305
> At2g33350
Length=440
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 7 EESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLK 48
E+ST+ + +L +K K + Y K + +++ NKK+K A K
Sbjct 326 EDSTLNKVGKLSPEQRKEKIRRYMKKRNERNFNKKIKYACRK 367
Lambda K H
0.313 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40