bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2596_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
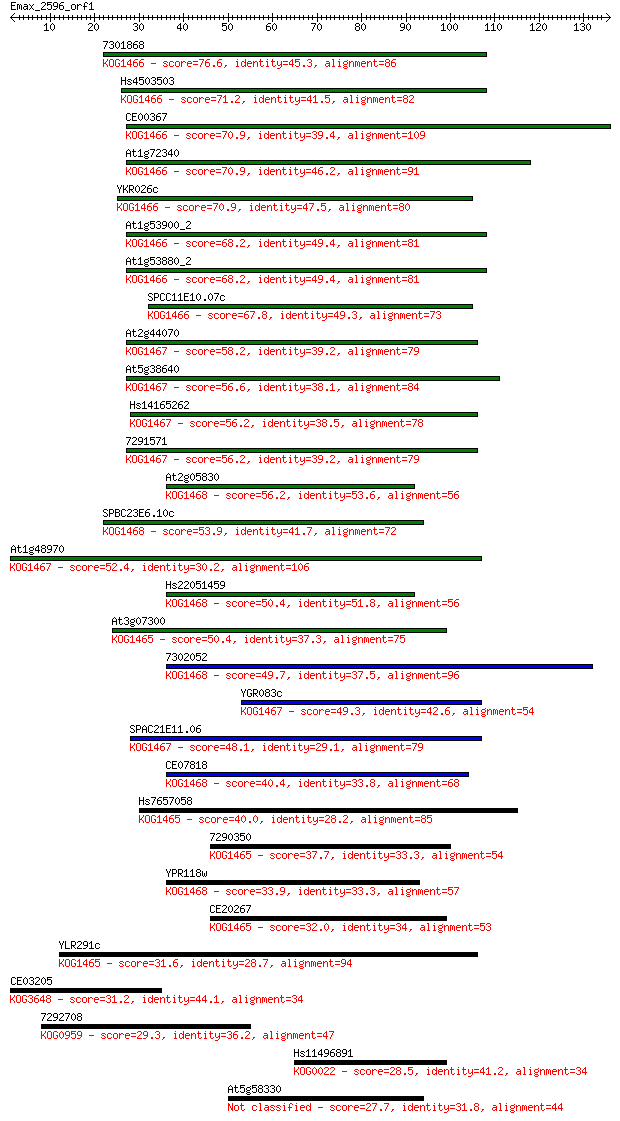
7301868 76.6 1e-14
Hs4503503 71.2 6e-13
CE00367 70.9 6e-13
At1g72340 70.9 7e-13
YKR026c 70.9 7e-13
At1g53900_2 68.2 4e-12
At1g53880_2 68.2 4e-12
SPCC11E10.07c 67.8 6e-12
At2g44070 58.2 5e-09
At5g38640 56.6 1e-08
Hs14165262 56.2 2e-08
7291571 56.2 2e-08
At2g05830 56.2 2e-08
SPBC23E6.10c 53.9 7e-08
At1g48970 52.4 2e-07
Hs22051459 50.4 1e-06
At3g07300 50.4 1e-06
7302052 49.7 1e-06
YGR083c 49.3 2e-06
SPAC21E11.06 48.1 4e-06
CE07818 40.4 0.001
Hs7657058 40.0 0.001
7290350 37.7 0.007
YPR118w 33.9 0.084
CE20267 32.0 0.30
YLR291c 31.6 0.46
CE03205 31.2 0.57
7292708 29.3 2.0
Hs11496891 28.5 3.5
At5g58330 27.7 6.3
> 7301868
Length=306
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/86 (45%), Positives = 53/86 (61%), Gaps = 0/86 (0%)
Query 22 NDEDRLLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALV 81
N + ++ L A GI +L S +ME VD V+VGAEAV E+GGIINR+GT T+ L
Sbjct 166 NSGEEMVKDLHAAGIDCTLILDSATGYVMESVDFVLVGAEAVVESGGIINRIGTYTMGLC 225
Query 82 AAQRCIPFYVVCEACKFGRAISLDNK 107
A + PFYV+ E+ KF R L+ +
Sbjct 226 AREMKKPFYVLAESFKFSRLYPLNQR 251
> Hs4503503
Length=305
Score = 71.2 bits (173), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 52/82 (63%), Gaps = 0/82 (0%)
Query 26 RLLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQR 85
++ +L +P ++ + V +ME+ DLV+VGAE V ENGGIIN++GT +A+ A +
Sbjct 163 KMAKALCHLNVPVTVVLDAAVGYIMEKADLVIVGAEGVVENGGIINKIGTNQMAVCAKAQ 222
Query 86 CIPFYVVCEACKFGRAISLDNK 107
PFYVV E+ KF R L+ +
Sbjct 223 NKPFYVVAESFKFVRLFPLNQQ 244
> CE00367
Length=305
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 62/112 (55%), Gaps = 6/112 (5%)
Query 27 LLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRC 86
+ L G+P +L S V +MER+ V+VGAE V E GGIIN++GT V ++A R
Sbjct 168 VFEELKKNGVPTTLVLDSCVGYVMERIQAVLVGAEGVMETGGIINKIGTVNVCIIAKSRH 227
Query 87 IPFYVVCEACKFGRAISLDNKAS--SLFSMQPAQPQRDSLIGDCWEP-LDYT 135
+P YV E KF R L N+A F + + +R++L + P +DYT
Sbjct 228 VPVYVCAETIKFVREFPL-NQADIPQEFKYRTSVIERNNL--ELEHPDVDYT 276
> At1g72340
Length=362
Score = 70.9 bits (172), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 42/91 (46%), Positives = 50/91 (54%), Gaps = 5/91 (5%)
Query 27 LLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRC 86
L + LS IP L S VA M+ VD+V VGA+ V E+GGIIN +GT +ALVA
Sbjct 224 LSSELSKLDIPVKLLLDSAVAYSMDEVDMVFVGADGVVESGGIINMMGTYQIALVAHSMN 283
Query 87 IPFYVVCEACKFGRAISLDNKASSLFSMQPA 117
P YV E+ KF R LD K M PA
Sbjct 284 KPVYVAAESYKFARLYPLDQK-----DMAPA 309
> YKR026c
Length=305
Score = 70.9 bits (172), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 38/80 (47%), Positives = 51/80 (63%), Gaps = 0/80 (0%)
Query 25 DRLLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQ 84
++L L GIP +L S V +++++VD V VGAE VAE+GGIIN VGT +V ++A
Sbjct 163 NQLYTLLEQKGIPVTLIVDSAVGAVIDKVDKVFVGAEGVAESGGIINLVGTYSVGVLAHN 222
Query 85 RCIPFYVVCEACKFGRAISL 104
PFYVV E+ KF R L
Sbjct 223 ARKPFYVVTESHKFVRMFPL 242
> At1g53900_2
Length=402
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/82 (48%), Positives = 49/82 (59%), Gaps = 1/82 (1%)
Query 27 LLAS-LSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQR 85
LLA+ L+ IP L S VA M+ VD+V VGA+ V E+GGIIN +GT +ALVA
Sbjct 263 LLANELAKLDIPVKLLIDSAVAYSMDEVDMVFVGADGVVESGGIINMMGTYQIALVAQSM 322
Query 86 CIPFYVVCEACKFGRAISLDNK 107
P YV E+ KF R LD K
Sbjct 323 NKPVYVAAESYKFARLYPLDQK 344
> At1g53880_2
Length=402
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/82 (48%), Positives = 49/82 (59%), Gaps = 1/82 (1%)
Query 27 LLAS-LSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQR 85
LLA+ L+ IP L S VA M+ VD+V VGA+ V E+GGIIN +GT +ALVA
Sbjct 263 LLANELAKLDIPVKLLIDSAVAYSMDEVDMVFVGADGVVESGGIINMMGTYQIALVAQSM 322
Query 86 CIPFYVVCEACKFGRAISLDNK 107
P YV E+ KF R LD K
Sbjct 323 NKPVYVAAESYKFARLYPLDQK 344
> SPCC11E10.07c
Length=341
Score = 67.8 bits (164), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 46/73 (63%), Gaps = 1/73 (1%)
Query 32 SACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYV 91
+AC IP + S V+ M RVDLV+VGAE V ENGG+IN++GT +A+ A PFY
Sbjct 184 NAC-IPTCMVLDSAVSFTMNRVDLVLVGAEGVVENGGLINQIGTFQLAVFAKHAHKPFYA 242
Query 92 VCEACKFGRAISL 104
V E+ KF R L
Sbjct 243 VAESHKFVRMFPL 255
> At2g44070
Length=307
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 46/79 (58%), Gaps = 0/79 (0%)
Query 27 LLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRC 86
LL L GI + + ++ ++ +M +V V +GA +V NG + +RVGTA VA+VA
Sbjct 144 LLRRLIKRGINCTYTHITAISYIMHQVTKVFLGASSVFSNGTVYSRVGTACVAMVANAFR 203
Query 87 IPFYVVCEACKFGRAISLD 105
+P V CEA KF + LD
Sbjct 204 VPVLVCCEAYKFHEKVQLD 222
> At5g38640
Length=642
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/84 (38%), Positives = 48/84 (57%), Gaps = 0/84 (0%)
Query 27 LLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRC 86
LL L GI + + ++ ++ +M V V +GA +V NG + +RVGTA VA+VA
Sbjct 486 LLRRLIKRGINCTYTHINAISYIMHEVTKVFLGASSVLSNGTVYSRVGTACVAMVANAFR 545
Query 87 IPFYVVCEACKFGRAISLDNKASS 110
+P V CEA KF + LD+ S+
Sbjct 546 VPVLVCCEAYKFHERVQLDSICSN 569
> Hs14165262
Length=523
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 28 LASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCI 87
L SL G+PAS + + ++ V V++GA A+ NG +++RVGTA +ALVA +
Sbjct 373 LRSLVHAGVPASYLLIPAASYVLPEVSKVLLGAHALLANGSVMSRVGTAQLALVARAHNV 432
Query 88 PFYVVCEACKFGRAISLD 105
P V CE KF + D
Sbjct 433 PVLVCCETYKFCERVQTD 450
> 7291571
Length=626
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 41/79 (51%), Gaps = 0/79 (0%)
Query 27 LLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRC 86
LL L A GIP + ++ V +M V++GA A+ NG ++ R GTA VALVA
Sbjct 476 LLRRLHATGIPCTYVLINAVGYVMAEATKVLLGAHALLANGYVMARTGTAQVALVANAHN 535
Query 87 IPFYVVCEACKFGRAISLD 105
+P V CE KF D
Sbjct 536 VPVLVCCETHKFSERFQTD 554
> At2g05830
Length=365
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/58 (51%), Positives = 40/58 (68%), Gaps = 2/58 (3%)
Query 36 IPASLSSVSLVASLME--RVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYV 91
IPA+L + S A+LM+ RVD V+VGA+ VA NG N++GT ++AL A IPFYV
Sbjct 226 IPATLIADSAAAALMKDGRVDGVIVGADRVASNGDTANKIGTYSLALCAKHHGIPFYV 283
> SPBC23E6.10c
Length=359
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Query 22 NDEDRLLA-SLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVAL 80
N RL A L IPA+L + S VAS+M ++D +VVGA+ V NG N++GT +A+
Sbjct 195 NQGSRLTAFELVHDKIPATLVTDSTVASIMHKIDAIVVGADRVTRNGDTANKIGTYNLAI 254
Query 81 VAAQRCIPFYVVC 93
+A PF V
Sbjct 255 LAKHFGKPFIVAA 267
> At1g48970
Length=756
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 53/106 (50%), Gaps = 9/106 (8%)
Query 1 RFIVIITQQETLNTHGQQTITNDEDRLLASLSACGIPASLSSVSLVASLMERVDLVVVGA 60
+F V+I N GQ +LL L G+ + + ++ ++ +M V +GA
Sbjct 577 KFRVVIVDSRP-NLEGQ--------KLLRRLVTRGLDCTYTHINAISYIMREATRVFLGA 627
Query 61 EAVAENGGIINRVGTATVALVAAQRCIPFYVVCEACKFGRAISLDN 106
++ NG + RVGT+ +A+VA +P V CEA KF + LD+
Sbjct 628 SSIYSNGTLYARVGTSCIAMVANAFSVPVIVCCEAYKFHERVLLDS 673
> Hs22051459
Length=369
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 37/58 (63%), Gaps = 2/58 (3%)
Query 36 IPASLSSVSLVASLMER--VDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYV 91
IPA+L + S+VA+ M V VVVGA+ V NG N+VGT +A+VA IPFYV
Sbjct 221 IPATLITDSMVAAAMAHRGVSAVVVGADRVVANGDTANKVGTYQLAIVAKHHGIPFYV 278
> At3g07300
Length=384
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 45/76 (59%), Gaps = 1/76 (1%)
Query 24 EDRLLAS-LSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVA 82
+ LLA L A G+ ++ + S V +++ RV++V++GA AV NGG+I VG AL A
Sbjct 247 QGHLLAKELVARGLQTTVITDSAVFAMISRVNMVIIGAHAVMANGGVIGPVGVNMAALAA 306
Query 83 AQRCIPFYVVCEACKF 98
+ +PF V+ + K
Sbjct 307 QKHAVPFVVLAGSHKL 322
> 7302052
Length=364
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 52/105 (49%), Gaps = 11/105 (10%)
Query 36 IPASLSSVSLVASLM--ERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYVVC 93
PA+L S+VA+L+ + V VVVGA+ VA NG N++GT +A+VA +PFYV
Sbjct 227 FPATLVLDSMVAALLRAKNVAAVVVGADRVASNGDTANKIGTYQIAVVAKHHDVPFYVAA 286
Query 94 E------ACKFGRAISLDNKASSLFSMQPAQPQRDSLIG-DCWEP 131
A G I ++ + M R + G +CW P
Sbjct 287 PLTSIDLAIPGGDHIIIEERPDR--EMTHVGEHRIAAPGINCWNP 329
> YGR083c
Length=651
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 53 VDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYVVCEACKFGRAISLDN 106
VD V +GA ++ NG + +R GTA +A+ A +R IP V CE+ KF + + LD+
Sbjct 467 VDYVFLGAHSILSNGFLYSRAGTAMLAMSAKRRNIPVLVCCESLKFSQRVQLDS 520
> SPAC21E11.06
Length=467
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 28 LASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCI 87
L L+ GI + +S ++ +M+ V + +G A+ NG + +R GT+ ++L+ + +
Sbjct 311 LKLLTEHGIECTYVMISALSYIMQEVTKIFLGGHAMLSNGALYSRAGTSLISLLGHESNV 370
Query 88 PFYVVCEACKFGRAISLDN 106
P CE+ KF I LD+
Sbjct 371 PVIACCESYKFTERIQLDS 389
> CE07818
Length=366
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query 36 IPASLSSVSLVASLME--RVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYVVC 93
+P L + S+ A M+ +VD V+ GA+ VA NG N++GT +A++ I FY V
Sbjct 233 VPFKLITDSMAAWAMKNHQVDCVLTGADNVARNGDTANKIGTYMLAVLCKHHNINFYPVV 292
Query 94 EACKFGRAIS 103
+ I+
Sbjct 293 PFTTINKNIA 302
> Hs7657058
Length=351
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 2/85 (2%)
Query 30 SLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPF 89
+LS GI ++ + + + ++M RV+ V++G + + NG + GT T+AL A P
Sbjct 206 NLSKAGIETTVMTDAAIFAVMSRVNKVIIGTKTILANGALRAVTGTHTLALAAKHHSTPL 265
Query 90 YVVCEACKFGRAISLDNKASSLFSM 114
+VC A F + N+ S
Sbjct 266 -IVC-APMFKLSPQFPNEEDSFHKF 288
> 7290350
Length=352
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 46 VASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYVVCEACKFG 99
+ ++M RV+ V++G +V NGG+ G TVAL A +P V+ K
Sbjct 224 IFAMMSRVNKVIIGTHSVLANGGLRAACGAYTVALAAKHYSVPVIVLAPMYKLS 277
> YPR118w
Length=411
Score = 33.9 bits (76), Expect = 0.084, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 33/61 (54%), Gaps = 4/61 (6%)
Query 36 IPASLSSVSLVASLMER----VDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYV 91
IP++L + S +A + + VGA+ + NG N++GT +A++ Q I F+V
Sbjct 251 IPSTLITDSSIAYRIRTSPIPIKAAFVGADRIVRNGDTANKIGTLQLAVICKQFGIKFFV 310
Query 92 V 92
V
Sbjct 311 V 311
> CE20267
Length=340
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 46 VASLMERVDLVVVGAEAVAENGGIINRVGTATVALVAAQRCIPFYVVCEACKF 98
VAS M VV+ A A +G + G +AL+A + C+P YV+ K
Sbjct 211 VASKMPEASKVVLVAAAYFPDGSCLVPAGGHQIALLAKRHCVPVYVLAPFYKL 263
> YLR291c
Length=381
Score = 31.6 bits (70), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 39/95 (41%), Gaps = 1/95 (1%)
Query 12 LNTHGQQTITNDEDRLLASLSACGIPASLSSVSLVASLMERVDLVVVGAEAVAENGGII- 70
L T G T + L+ I + S V +LM RV V++G +AV NGG I
Sbjct 214 LVTEGFPNNTKNAHEFAKKLAQHNIETLVVPDSAVFALMSRVGKVIIGTKAVFVNGGTIS 273
Query 71 NRVGTATVALVAAQRCIPFYVVCEACKFGRAISLD 105
+ G ++V A + P + V K D
Sbjct 274 SNSGVSSVCECAREFRTPVFAVAGLYKLSPLYPFD 308
> CE03205
Length=1149
Score = 31.2 bits (69), Expect = 0.57, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 1 RFIVIITQQETLNTHGQQTITNDEDRLLASLSAC 34
RF +I+ L T QQ + ND D+ LAS AC
Sbjct 3 RFPLILASVCWLTTAQQQNVANDPDKKLASFDAC 36
> 7292708
Length=1077
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 8 QQETLNTHGQQTITNDE---DRLLASLSACGIPASLSSVSLVASLMERVD 54
++ +++ QQ + +DE D+LLASL+ G P + + SL E VD
Sbjct 190 ERSAVDSEFQQILQDDETRRDQLLASLATKGFPHGTFAWGNMKSLKENVD 239
> Hs11496891
Length=374
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 16/35 (45%), Gaps = 1/35 (2%)
Query 65 ENGGIINRVGTATVALVAAQRCIPFYVV-CEACKF 98
E GI+ VG L A IP Y+ C CKF
Sbjct 68 EGAGIVESVGEGVTKLKAGDTVIPLYIPQCGECKF 102
> At5g58330
Length=442
Score = 27.7 bits (60), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 50 MERVDLVVVGAEAVAENGGIINRVGTATVA-LVAAQRCIPFYVVC 93
MER DL+ + + AE G +N+ + V LV C ++C
Sbjct 190 MERADLLDINGQIFAEQGKALNKAASPNVKVLVVGNPCNTNALIC 234
Lambda K H
0.320 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40