bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2580_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
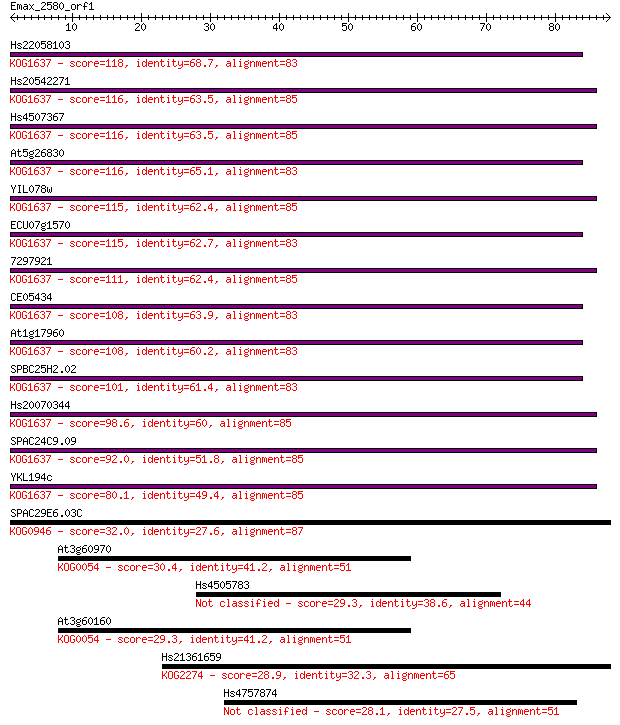
Hs22058103 118 2e-27
Hs20542271 116 1e-26
Hs4507367 116 1e-26
At5g26830 116 1e-26
YIL078w 115 2e-26
ECU07g1570 115 3e-26
7297921 111 3e-25
CE05434 108 2e-24
At1g17960 108 3e-24
SPBC25H2.02 101 4e-22
Hs20070344 98.6 3e-21
SPAC24C9.09 92.0 2e-19
YKL194c 80.1 1e-15
SPAC29E6.03C 32.0 0.31
At3g60970 30.4 0.89
Hs4505783 29.3 1.9
At3g60160 29.3 2.0
Hs21361659 28.9 2.1
Hs4757874 28.1 4.0
> Hs22058103
Length=707
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 57/83 (68%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFCT+EQ+EEE L+FL VY FGFSF L LST
Sbjct 425 HRNELSGTLSGLTRVRRFQQDDAHIFCTVEQIEEEIKGCLQFLQSVYSTFGFSFQLNLST 484
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP +G IE WN+AE LQN+L
Sbjct 485 RPENFLGEIEMWNEAEKQLQNSL 507
> Hs20542271
Length=723
Score = 116 bits (290), Expect = 1e-26, Method: Composition-based stats.
Identities = 54/85 (63%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC +EQ+E+E L+FL VY FGFSF L LST
Sbjct 441 HRNELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGFSFKLNLST 500
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IE W+QAE L+N+L +
Sbjct 501 RPEKFLGDIEVWDQAEKQLENSLNE 525
> Hs4507367
Length=712
Score = 116 bits (290), Expect = 1e-26, Method: Composition-based stats.
Identities = 54/85 (63%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC +EQ+E+E L+FL VY FGFSF L LST
Sbjct 429 HRNELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGFSFKLNLST 488
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IE W+QAE L+N+L +
Sbjct 489 RPEKFLGDIEVWDQAEKQLENSLNE 513
> At5g26830
Length=676
Score = 116 bits (290), Expect = 1e-26, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 67/83 (80%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+EASG+LSGLTRVRRFQQDDAHIFCT EQV+ E LEF+ YVY+ FGF++ L LST
Sbjct 402 HRNEASGALSGLTRVRRFQQDDAHIFCTTEQVKGEVQGVLEFIDYVYKVFGFTYELKLST 461
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP K +G +ETW++AE L+ A+
Sbjct 462 RPEKYLGDLETWDKAEADLKEAI 484
> YIL078w
Length=734
Score = 115 bits (288), Expect = 2e-26, Method: Composition-based stats.
Identities = 53/85 (62%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFCT +Q+E E + FL Y+Y FGF F + LST
Sbjct 448 HRNEFSGALSGLTRVRRFQQDDAHIFCTHDQIESEIENIFNFLQYIYGVFGFEFKMELST 507
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IETW+ AE+ L++AL+K
Sbjct 508 RPEKYVGKIETWDAAESKLESALKK 532
> ECU07g1570
Length=640
Score = 115 bits (287), Expect = 3e-26, Method: Composition-based stats.
Identities = 52/83 (62%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFCT +QV+EE LEFL +VY FGF F L LST
Sbjct 358 HRNELSGTLTGLTRVRRFQQDDAHIFCTKDQVKEEIKGCLEFLSFVYGVFGFRFELVLST 417
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP K +G ++ W++AE AL +A+
Sbjct 418 RPEKYLGSVDEWDRAEKALADAM 440
> 7297921
Length=690
Score = 111 bits (278), Expect = 3e-25, Method: Composition-based stats.
Identities = 53/85 (62%), Positives = 62/85 (72%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC EQ++ E LEFL YVY FGFSF L LST
Sbjct 410 HRNELSGALTGLTRVRRFQQDDAHIFCAPEQIKSEMKGCLEFLKYVYTIFGFSFQLVLST 469
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP +G +E WN AE AL +L +
Sbjct 470 RPDNYLGELEQWNDAEKALAESLNE 494
> CE05434
Length=725
Score = 108 bits (271), Expect = 2e-24, Method: Composition-based stats.
Identities = 53/84 (63%), Positives = 64/84 (76%), Gaps = 1/84 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQ-FGFSFSLALS 59
HR+E SG+L+GLTRVRRFQQDDAHIFC +Q+ EE L+FL Y YE+ FGF+F L LS
Sbjct 440 HRNEMSGALTGLTRVRRFQQDDAHIFCRQDQISEEIKQCLDFLEYAYEKVFGFTFKLNLS 499
Query 60 TRPLKAIGCIETWNQAETALQNAL 83
TRP +G IETW++AE L NAL
Sbjct 500 TRPEGFLGNIETWDKAEADLTNAL 523
> At1g17960
Length=458
Score = 108 bits (269), Expect = 3e-24, Method: Composition-based stats.
Identities = 50/83 (60%), Positives = 62/83 (74%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L G+TRVRRF QDDAHIFC ++QVEEE L+F+ YVY FGF++ L LST
Sbjct 181 HRNEDSGALGGMTRVRRFVQDDAHIFCRVDQVEEEVKGVLDFIDYVYRIFGFTYELTLST 240
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP IG +ETW +AE L+ AL
Sbjct 241 RPKDHIGDLETWAKAENDLEKAL 263
> SPBC25H2.02
Length=703
Score = 101 bits (251), Expect = 4e-22, Method: Composition-based stats.
Identities = 51/84 (60%), Positives = 61/84 (72%), Gaps = 1/84 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFCT +QV E +FL VY FGF+F L LST
Sbjct 409 HRNEFSGALSGLTRVRRFQQDDAHIFCTPDQVRSEIEGCFDFLKEVYGTFGFTFHLELST 468
Query 61 RP-LKAIGCIETWNQAETALQNAL 83
RP K +G + TW++AE L+ AL
Sbjct 469 RPEEKYLGDLATWDKAEAQLKAAL 492
> Hs20070344
Length=718
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 51/85 (60%), Positives = 60/85 (70%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR EASG L GLTR+R FQQDDAHIFCT +Q+E E S L+FL VY GFSF LALST
Sbjct 437 HRAEASGGLGGLTRLRCFQQDDAHIFCTTDQLEAEIQSCLDFLRSVYAVLGFSFRLALST 496
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP +G W+QAE L+ AL++
Sbjct 497 RPSGFLGDPCLWDQAEQVLKQALKE 521
> SPAC24C9.09
Length=473
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 61/86 (70%), Gaps = 1/86 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFS-FSLALS 59
HR+EASG+LSGLTR+R F QDD HIFC+ E +++E + L F+ VY G + L LS
Sbjct 174 HRNEASGALSGLTRLRCFHQDDGHIFCSPESIKDEIKNTLTFVKQVYSLLGMNKLKLYLS 233
Query 60 TRPLKAIGCIETWNQAETALQNALRK 85
TRP + IG ++TWN+AE L+ AL++
Sbjct 234 TRPEEHIGSLDTWNEAENGLREALQE 259
> YKL194c
Length=462
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 42/95 (44%), Positives = 59/95 (62%), Gaps = 10/95 (10%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQ-FGF------- 52
HR+EASG+LSGLTR+R+F QDD HIFCT QV+ E ++L+ + VY + F F
Sbjct 161 HRNEASGALSGLTRLRKFHQDDGHIFCTPSQVKSEIFNSLKLIDIVYNKIFPFVKGGSGA 220
Query 53 --SFSLALSTRPLKAIGCIETWNQAETALQNALRK 85
++ + STRP IG ++ WN AE L+ L +
Sbjct 221 ESNYFINFSTRPDHFIGDLKVWNHAEQVLKEILEE 255
> SPAC29E6.03C
Length=1044
Score = 32.0 bits (71), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 7/87 (8%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
H D++ S +GL +F + +I C L+ + + FYV FS LS
Sbjct 93 HDDDSRASDTGLWIADQFILNQDNIQCLLQSISHKD-------FYVRLYSVELFSAILSC 145
Query 61 RPLKAIGCIETWNQAETALQNALRKHI 87
RP + C++T+ A +++ LR I
Sbjct 146 RPTELKDCLQTFPSAISSIMVPLRDSI 172
> At3g60970
Length=1037
Score = 30.4 bits (67), Expect = 0.89, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 8 SLSGLTRVRRFQQDDAHIFCTLEQVEEEG------ASALEFLFY---VYEQFGFSFSLAL 58
SL+G T +R F Q D I L ++ ASA+E+L + + F F+FSL L
Sbjct 650 SLAGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRLNLLSHFVFAFSLVL 709
> Hs4505783
Length=1093
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 28 TLEQVEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGCIET 71
T +QVE + L Y Q G + L LS RP + IGC+ T
Sbjct 519 TPQQVEPIQIWPQQELVKAYLQLGINEKLGLSGRPDRPIGCLGT 562
> At3g60160
Length=1490
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 8 SLSGLTRVRRFQQDDAHIFCTLEQVEEEG------ASALEFLFY---VYEQFGFSFSLAL 58
SL+G T +R F Q D I L ++ ASA+E+L + + F F+FSL L
Sbjct 1103 SLAGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRLNLLSHFVFAFSLVL 1162
> Hs21361659
Length=1041
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 23 AHIFCTLEQVEEEGASALEFLFYVYEQFGFSF--SLALSTRPLKAIGCIETWNQAETALQ 80
AH+ C +E++E+ A L +F V +QF +F +L + P G +A TAL
Sbjct 219 AHMICNMEELEKGAAKVL--IFPVVQQFTEAFVQALQIPDGPTSDSGFKMEVLKAVTALV 276
Query 81 NALRKHI 87
KH+
Sbjct 277 KNFPKHM 283
> Hs4757874
Length=318
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 32 VEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGCIETWNQAETALQNA 82
VE G +++ L + GF +S RP+K + C++ + AL++A
Sbjct 235 VESCGEGSMKVLEKRLKDMGFQYSCINDYRPVKLLQCVDHSTHPDCALKSA 285
Lambda K H
0.322 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187582654
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40