bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2563_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
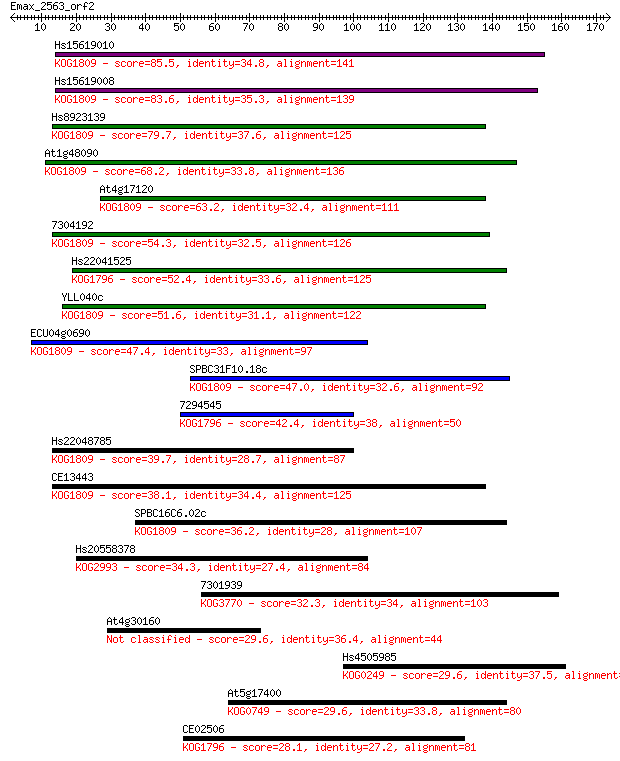
Hs15619010 85.5 4e-17
Hs15619008 83.6 2e-16
Hs8923139 79.7 3e-15
At1g48090 68.2 7e-12
At4g17120 63.2 2e-10
7304192 54.3 1e-07
Hs22041525 52.4 4e-07
YLL040c 51.6 8e-07
ECU04g0690 47.4 2e-05
SPBC31F10.18c 47.0 2e-05
7294545 42.4 5e-04
Hs22048785 39.7 0.003
CE13443 38.1 0.009
SPBC16C6.02c 36.2 0.029
Hs20558378 34.3 0.13
7301939 32.3 0.48
At4g30160 29.6 2.8
Hs4505985 29.6 3.1
At5g17400 29.6 3.3
CE02506 28.1 9.2
> Hs15619010
Length=3174
Score = 85.5 bits (210), Expect = 4e-17, Method: Composition-based stats.
Identities = 49/142 (34%), Positives = 69/142 (48%), Gaps = 7/142 (4%)
Query 14 DSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMWS-LTNIVTKPIEGAQREGVGGFFKG 72
D +Y + + + A R+G GK + G S +T IVTKPI+GAQ+ G GFFKG
Sbjct 2938 DEDYQQKRREAMNKQPAGFREGITRGGKGLVSGFVSGITGIVTKPIKGAQKGGAAGFFKG 2997
Query 73 IGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKFRNERRRKPRMLRELGEIRPY 132
+GKG+VG++ +P + S +GIK E R PR E G IRPY
Sbjct 2998 VGKGLVGAVARPTGGIIDMASSTFQGIKRATETS------EVESLRPPRFFNEDGVIRPY 3051
Query 133 DETEATLRECLGLAVTRRLQKF 154
+ T + L + R K+
Sbjct 3052 RLRDGTGNQMLQVMENGRFAKY 3073
> Hs15619008
Length=3095
Score = 83.6 bits (205), Expect = 2e-16, Method: Composition-based stats.
Identities = 49/140 (35%), Positives = 70/140 (50%), Gaps = 12/140 (8%)
Query 14 DSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMWS-LTNIVTKPIEGAQREGVGGFFKG 72
D +Y + + + A R+G GK + G S +T IVTKPI+GAQ+ G GFFKG
Sbjct 2938 DEDYQQKRREAMNKQPAGFREGITRGGKGLVSGFVSGITGIVTKPIKGAQKGGAAGFFKG 2997
Query 73 IGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKFRNERRRKPRMLRELGEIRPY 132
+GKG+VG++ +P + S +GIK E R PR E G IRPY
Sbjct 2998 VGKGLVGAVARPTGGIIDMASSTFQGIKRATETS------EVESLRPPRFFNEDGVIRPY 3051
Query 133 DETEATLRECLGLAVTRRLQ 152
LR+ G + +++Q
Sbjct 3052 -----RLRDGTGNQMLQKIQ 3066
> Hs8923139
Length=442
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 68/128 (53%), Gaps = 10/128 (7%)
Query 13 FDSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMWS-LTNIVTKPIEGAQREGVGGFFK 71
D EY + + E R D GK G+ +T I+TKP+EGA++EG GFFK
Sbjct 183 MDKEYQQKRREELSRQPRDFGDSLARGGKGFLRGVVGGVTGIITKPVEGAKKEGAAGFFK 242
Query 72 GIGKGIVGSLVKPLDKVGQAVSDVTRGIK--AEVSRPIGAYKFRNERRRKPRMLRELGEI 129
GIGKG+VG++ +P + S +GI+ AE + + + R PR++ E G I
Sbjct 243 GIGKGLVGAVARPTGGIVDMASSTFQGIQRAAESTEEVSSL-------RPPRLIHEDGII 295
Query 130 RPYDETEA 137
RPYD E+
Sbjct 296 RPYDRQES 303
> At1g48090
Length=4099
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 46/140 (32%), Positives = 70/140 (50%), Gaps = 5/140 (3%)
Query 11 FHFDSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMW-SLTNIVTKPIEGAQREGVGGF 69
D ++I QR+ + D G + +G++ +T I+TKP+EGA+ GV GF
Sbjct 3791 LSMDKKFIQSRQRQENKGVEDFGDIIREGGGALAKGLFRGVTGILTKPLEGAKSSGVEGF 3850
Query 70 FKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKFRNE---RRRKPRMLREL 126
G GKGI+G+ +P+ V +S T G A + I A +E RRR PR +
Sbjct 3851 VSGFGKGIIGAAAQPVSGVLDLLSKTTEGANA-MRMKIAAAITSDEQLLRRRLPRAVGAD 3909
Query 127 GEIRPYDETEATLRECLGLA 146
+RPY++ A + L LA
Sbjct 3910 SLLRPYNDYRAQGQVILQLA 3929
> At4g17120
Length=1661
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/114 (31%), Positives = 58/114 (50%), Gaps = 3/114 (2%)
Query 27 RNTASMRDGFLSAGKNIGEGM-WSLTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPL 85
R + D + + + +G+ + ++ +VTKP+E A+ G+ GF G+G+ +G +V+P+
Sbjct 1536 RRITGVGDAIVQGTEALAQGVAFGVSGVVTKPVESARENGILGFAHGVGRAFLGFIVQPV 1595
Query 86 DKVGQAVSDVTRGIKAEVSRPIGAYKFRN--ERRRKPRMLRELGEIRPYDETEA 137
S GI A SR + R ER R PR + G +R YDE EA
Sbjct 1596 SGALDFFSLTVDGIGASCSRCLEVLSNRTALERIRNPRAVHADGILREYDEKEA 1649
> 7304192
Length=3242
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 41/127 (32%), Positives = 60/127 (47%), Gaps = 6/127 (4%)
Query 13 FDSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMWS-LTNIVTKPIEGAQREGVGGFFK 71
FD +Y + ++ + +G + K + G +T +VTKP+ GA+ GV GFFK
Sbjct 2989 FDEDYQKKRRQGIQNKPKNFHEGLARSSKGLVMGFVDGVTGVVTKPVTGARDNGVEGFFK 3048
Query 72 GIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKFRNERRRKPRMLRELGEIRP 131
G+GKG +G + +P V D G V R A + +R R PR +RP
Sbjct 3049 GLGKGAIGLVARP----TAGVVDFASGSFEAVKRAADASE-DVKRMRPPRFQHYDFVLRP 3103
Query 132 YDETEAT 138
Y EAT
Sbjct 3104 YCLMEAT 3110
> Hs22041525
Length=1231
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 70/131 (53%), Gaps = 10/131 (7%)
Query 19 NRSQRERGRNTASMRDGFLSAG-KNIGEGMWS-LTNIVTKPIEGAQREG-VGGFFKGIGK 75
++S+RE R A+ L AG + G+ LT+++T +EG + EG V GF G+GK
Sbjct 971 HQSEREYIRYHAATSGEHLVAGIHGLAHGIIGGLTSVITSTVEGVKTEGGVSGFISGLGK 1030
Query 76 GIVGSLVKPLDKVGQAVSDVTRGIK--AEVSRPIGAYKFRNERRRKPRMLRELGEIRP-Y 132
G+VG++ KP+ S+ + ++ A +S P + + +R RKPR + P Y
Sbjct 1031 GLVGTVTKPVAGALDFASETAQAVRDTATLSGP----RTQAQRVRKPRCCTGPQGLLPRY 1086
Query 133 DETEATLRECL 143
E++A +E L
Sbjct 1087 SESQAEGQEQL 1097
> YLL040c
Length=3144
Score = 51.6 bits (122), Expect = 8e-07, Method: Composition-based stats.
Identities = 38/122 (31%), Positives = 57/122 (46%), Gaps = 12/122 (9%)
Query 16 EYINRSQRERGRNTASMRDGFLSAGKNIGEGMWSLTNIVTKPIEGAQREGVGGFFKGIGK 75
+ IN++ R N+A S +G G L+ I P + Q+EG GF KG+GK
Sbjct 2925 QRINKNNRNALANSAQ------SFASTLGSG---LSGIALDPYKAMQKEGAAGFLKGLGK 2975
Query 76 GIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKFRNERRRKPRMLRELGEIRPYDET 135
GIVG K S++++G+K+ + + R R PR + I+PYD
Sbjct 2976 GIVGLPTKTAIGFLDLTSNLSQGVKSTTTV---LDMQKGCRVRLPRYVDHDQIIKPYDLR 3032
Query 136 EA 137
EA
Sbjct 3033 EA 3034
> ECU04g0690
Length=2371
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 14/100 (14%)
Query 7 FAFNFHFDSEYINRSQRERGRNTASMR---DGFLSAGKNIGEGMWSLTNIVTKPIEGAQR 63
+A + H + RS+ +G + +R D F S + I I T PIEGA
Sbjct 2164 YACDVHL---LVPRSKHSKGSVVSILRGTGDLFDSITRGIA-------GIATSPIEGAS- 2212
Query 64 EGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEV 103
+GV G KG+GKGI+G+ +P+ +V V+ ++ IK +
Sbjct 2213 QGVTGVVKGLGKGILGAFTRPIVEVADLVTGISDTIKVSM 2252
> SPBC31F10.18c
Length=600
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 49/93 (52%), Gaps = 10/93 (10%)
Query 53 IVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYKF 112
+ +PI GA+R G+ G KG+GKG+VG KPL + S ++ G + + +
Sbjct 405 LALQPIIGARRNGLPGLVKGLGKGLVGFTTKPLVGLFDFASSISEGARNTTT----VFDE 460
Query 113 RN-ERRRKPRMLRELGEIRPYDETEATLRECLG 144
R+ E+ R R++ + G + P+ LRE LG
Sbjct 461 RHIEKLRLSRLMSDDGVVYPFQ-----LREALG 488
> 7294545
Length=1902
Score = 42.4 bits (98), Expect = 5e-04, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 50 LTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGI 99
+T+IV +GA +GV GF G+GKG+VG++ KP+ V S+ +
Sbjct 1788 VTSIVRHTYDGATSDGVPGFLSGLGKGLVGTVTKPIIGVLDLASETASAV 1837
> Hs22048785
Length=1687
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 42/97 (43%), Gaps = 10/97 (10%)
Query 13 FDSEYINRSQRERGRNTASMRDGFLSAGKNIGEGMW-SLTNIVTKPIEGAQREG------ 65
D E+ NR + R + S+ +G +G + ++ IV +P++ Q+
Sbjct 1393 LDEEHYNRQEEWRRQLPESLGEGLRQGLSRLGISLLGAIAGIVDQPMQNFQKTSEAQASA 1452
Query 66 ---VGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGI 99
G G+GKGI+G KP+ + VS GI
Sbjct 1453 GHKAKGVISGVGKGIMGVFTKPIGGAAELVSQTGYGI 1489
> CE13443
Length=3212
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 67/128 (52%), Gaps = 10/128 (7%)
Query 13 FDSEYINRSQRERGRNTASMRDGFLSAGKNIG-EGMWSLTNIVTKPIEGAQREGVGGFFK 71
FD +Y+ + Q + R S +G K +G + +T +VTKPIEGA++EG GF K
Sbjct 2976 FDDDYMKKRQEDLNRKPQSFGEGMARGLKGLGMGVVGGITGVVTKPIEGAKQEGGFGFVK 3035
Query 72 GIGKGIVGSLVKPLDKVGQAVSDVTRGIKAE--VSRPIGAYKFRNERRRKPRMLRELGEI 129
G+GKG++G + +P+ V S ++A +R G R PR+LRE +
Sbjct 3036 GVGKGLIGVVTRPVSGVVDFASGTMNSVRAVAGTNREAGPL-------RPPRVLREDKIV 3088
Query 130 RPYDETEA 137
+PY +A
Sbjct 3089 KPYSSGDA 3096
> SPBC16C6.02c
Length=3131
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 7/111 (6%)
Query 37 LSAGKN-IGEGMWSLTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDV 95
++AG N + M S + KP + G F KG GKG++G KP + S+V
Sbjct 2794 VTAGANSFYDSMSSGFKGLKKPFTDPKNNSAGKFLKGFGKGMLGLATKPAIGLLDMTSNV 2853
Query 96 TRGIKAEVSRPIGAYKFRNERRRKPRMLRELGEI---RPYDETEATLRECL 143
+ GI+ ++ R PR + G I +PY+ + CL
Sbjct 2854 SEGIRNSTDVRTNP---EIDKVRVPRYVEFGGLIVPFKPYESLGKYMLSCL 2901
> Hs20558378
Length=1938
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 44/87 (50%), Gaps = 3/87 (3%)
Query 20 RSQRE--RGRNTASMRDGFLSAGKNIGEGMW-SLTNIVTKPIEGAQREGVGGFFKGIGKG 76
RS R RG+ A +R+G A + EG+ + I G +++G+ G G+ +
Sbjct 1833 RSARRLRRGQQPADLREGVAKAYDTVREGILDTAQTICDVASRGHEQKGLTGAVGGVIRQ 1892
Query 77 IVGSLVKPLDKVGQAVSDVTRGIKAEV 103
+ ++VKPL +A S + G++ ++
Sbjct 1893 LPPTVVKPLILATEATSSLLGGMRNQI 1919
> 7301939
Length=638
Score = 32.3 bits (72), Expect = 0.48, Method: Composition-based stats.
Identities = 35/107 (32%), Positives = 45/107 (42%), Gaps = 11/107 (10%)
Query 56 KPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGAYK---F 112
K I G G+G F G G + S+ L K QA S V+ I E+SR Y
Sbjct 2 KLIRGTTLLGIGILFVGCGAFSLPSVYDVLSKDQQATSFVSASIAEEISREYLKYHRTGI 61
Query 113 RNERRRKPRMLRELG-EIRPYDETEATLRECLGLAVTRRLQKFVTVL 158
ER LR+LG +IR +A E + +T Q FV L
Sbjct 62 ETER------LRQLGKDIRSSHSKKAIFTESMA-DLTSTDQFFVCTL 101
> At4g30160
Length=974
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 6/47 (12%)
Query 29 TASMRD---GFLSAGKNIGEGMWSLTNIVTKPIEGAQREGVGGFFKG 72
+ SMRD F AG+ G +W + N + PI + +G FF G
Sbjct 2 SVSMRDLDPAFQGAGQKAGIEIWRIENFIPTPI---PKSSIGKFFTG 45
> Hs4505985
Length=1257
Score = 29.6 bits (65), Expect = 3.1, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 36/71 (50%), Gaps = 18/71 (25%)
Query 97 RGIKAEVSRPIGAYKFRNERRRKPRMLRELGEIRPYDETEATLRECLGLA-------VTR 149
+GIK+ + R G ++ K R LG++R + ETEA +E LGL R
Sbjct 828 KGIKSSIGRLFG-------KKEKAR----LGQLRGFMETEAAAQESLGLGKLGTQAEKDR 876
Query 150 RLQKFVTVLKE 160
RL+K +L+E
Sbjct 877 RLKKKHELLEE 887
> At5g17400
Length=327
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 38/96 (39%), Gaps = 19/96 (19%)
Query 64 EGVGGFFKGIGKGIVG------------SLVKPLDKVGQA----VSDVTRGIKAEVSRPI 107
+G+ G ++G G IVG +KP+ VG ++ G S +
Sbjct 167 DGIKGLYRGFGVSIVGITLYRGMYFGMYDTIKPIVLVGSLEGNFLASFLLGWSITTSAGV 226
Query 108 GAYKFRNERRRKPRMLRELGEIRPYDETEATLRECL 143
AY F RR RM+ G+ Y T LRE L
Sbjct 227 IAYPFDTLRR---RMMLTSGQPVKYRNTIHALREIL 259
> CE02506
Length=2102
Score = 28.1 bits (61), Expect = 9.2, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 37/84 (44%), Gaps = 4/84 (4%)
Query 51 TNIVTKPIEGAQREG-VGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSRPIGA 109
T + T +++ G V G G+ G+V ++ KP+ V V +K E++ P
Sbjct 1901 TAMFTNVASESRKSGLVKGMVWGVATGVVDTVTKPVQGVFDFVEGTASAMK-ELAMPATG 1959
Query 110 YKFRNE--RRRKPRMLRELGEIRP 131
+ R R PR+ R L + P
Sbjct 1960 VRRATALCRVRIPRLCRNLYHLLP 1983
Lambda K H
0.322 0.141 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40