bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2487_orf1
Length=157
Score E
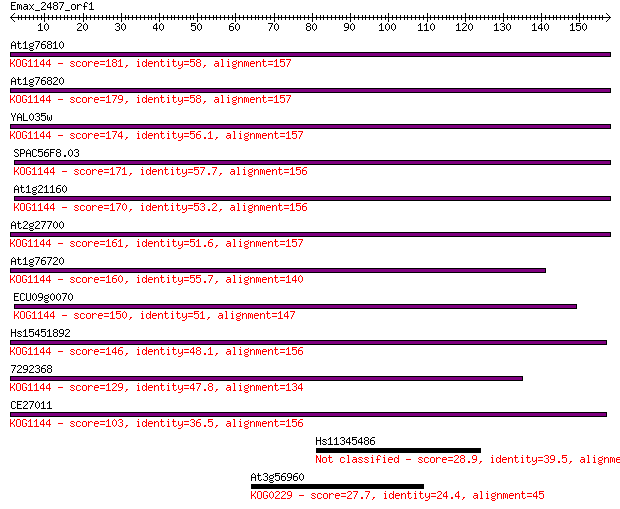
Sequences producing significant alignments: (Bits) Value
At1g76810 181 7e-46
At1g76820 179 1e-45
YAL035w 174 4e-44
SPAC56F8.03 171 5e-43
At1g21160 170 1e-42
At2g27700 161 5e-40
At1g76720 160 7e-40
ECU09g0070 150 1e-36
Hs15451892 146 1e-35
7292368 129 3e-30
CE27011 103 1e-22
Hs11345486 28.9 4.1
At3g56960 27.7 8.8
> At1g76810
Length=1280
Score = 181 bits (458), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 91/159 (57%), Positives = 117/159 (73%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ + ++A+ G
Sbjct 929 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHYKEIKAAQG 988
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGT+L VV D I I+ M+DM +VL +D +G GV+V ASTLGS
Sbjct 989 IKITAQGLEHAIAGTALHVVGPDDDIEAIKESAMEDMESVLSRIDKSGEGVYVQASTLGS 1048
Query 121 LEALLVYLEE--CKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL YL+ KIPV + IG V KKD+ +A VM E+
Sbjct 1049 LEALLEYLKSPAVKIPVSGIGIGPVHKKDVMKAGVMLER 1087
> At1g76820
Length=1146
Score = 179 bits (455), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 91/159 (57%), Positives = 115/159 (72%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ H ++A+ G
Sbjct 812 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHHKEIKAAQG 871
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGTSL VV D I ++ M+DM +VL +D +G GV+V STLGS
Sbjct 872 IKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVLSRIDKSGEGVYVQTSTLGS 931
Query 121 LEALLVYLE--ECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L+ IPV + IG V KKDI +A VM EK
Sbjct 932 LEALLEFLKTPAVNIPVSGIGIGPVHKKDIMKAGVMLEK 970
> YAL035w
Length=1002
Score = 174 bits (442), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 88/157 (56%), Positives = 118/157 (75%), Gaps = 0/157 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL G L EGD+IV+CG++GPI+T IR LLTPQPL+ELR+K EY+ H V+A++G
Sbjct 645 TTIDVILSNGYLREGDRIVLCGMNGPIVTNIRALLTPQPLRELRLKSEYVHHKEVKAALG 704
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI A L++AV+G+ L VV D ++ +VM D+ +L SVD+TG GV V ASTLGS
Sbjct 705 VKIAANDLEKAVSGSRLLVVGPEDDEDELMDDVMDDLTGLLDSVDTTGKGVVVQASTLGS 764
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L++ KIPV ++ +G V K+D+ +AS M EK
Sbjct 765 LEALLDFLKDMKIPVMSIGLGPVYKRDVMKASTMLEK 801
> SPAC56F8.03
Length=1079
Score = 171 bits (434), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 90/156 (57%), Positives = 118/156 (75%), Gaps = 0/156 (0%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
TIDVIL GVL+EGD+IV+CG+ GPIITT+R LLTPQPLKE+RVK Y+ H ++A+MGV
Sbjct 725 TIDVILSNGVLHEGDRIVLCGMGGPIITTVRALLTPQPLKEMRVKSAYVHHKEIKAAMGV 784
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSL 121
KI A L++AVAG+ L VV D D+ E+M+D+ +L +D++G GV V ASTLGSL
Sbjct 785 KICANDLEKAVAGSRLLVVGPDDDEEDLAEEIMEDLENLLGRIDTSGIGVSVQASTLGSL 844
Query 122 EALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
EALL +L++ KIPV +V+IG V KKD+ R + M EK
Sbjct 845 EALLEFLKQMKIPVASVNIGPVYKKDVMRCATMLEK 880
> At1g21160
Length=1088
Score = 170 bits (430), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 83/158 (52%), Positives = 115/158 (72%), Gaps = 2/158 (1%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
T+DV+LV GVL EGD+IV+CG GPI+TTIR+LLTP P+ E+RV G Y+ H V+A+ G+
Sbjct 732 TVDVVLVNGVLREGDQIVVCGSQGPIVTTIRSLLTPYPMNEMRVTGTYMPHREVKAAQGI 791
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSL 121
KI A GL+ A+AGT+L V+ + + + + M+D+ +V+ +D +G GV+V ASTLGSL
Sbjct 792 KIAAQGLEHAIAGTALHVIGPNEDMEEAKKNAMEDIESVMNRIDKSGEGVYVQASTLGSL 851
Query 122 EALLVYLE--ECKIPVFAVSIGTVQKKDIKRASVMREK 157
EALL +L+ + KIPV + IG V KKDI +A VM EK
Sbjct 852 EALLEFLKSSDVKIPVSGIGIGPVHKKDIMKAGVMLEK 889
> At2g27700
Length=479
Score = 161 bits (407), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 81/159 (50%), Positives = 112/159 (70%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EG +IV+CGL GPI+TTIR LLTP P+KEL V G ++ H ++A+
Sbjct 274 TTIDVVLVNGELHEGGQIVVCGLQGPIVTTIRALLTPHPIKELHVNGNHVHHEVIKAAEC 333
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+ I+A L+ + GT+L VV D I I+ VM+D+ +VL +D +G GV++ ASTLGS
Sbjct 334 INIIAKDLEHVIVGTALHVVGPDDDIEAIKELVMEDVNSVLSRIDKSGEGVYIQASTLGS 393
Query 121 LEALLVYLEE--CKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L+ K+PV + IG VQKKD+ +A VM E+
Sbjct 394 LEALLEFLKSPAVKLPVGGIGIGPVQKKDVMKAGVMLER 432
> At1g76720
Length=1224
Score = 160 bits (406), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 78/140 (55%), Positives = 101/140 (72%), Gaps = 3/140 (2%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ H ++A+ G
Sbjct 899 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHHKEIKAAQG 958
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGTSL VV D I ++ M+DM +VL +D +G GV+V STLGS
Sbjct 959 IKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVLSRIDKSGEGVYVQTSTLGS 1018
Query 121 LEALLVYLEECKIPVFAVSI 140
LEALL +L K P + +
Sbjct 1019 LEALLEFL---KTPAVNIPV 1035
> ECU09g0070
Length=659
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 75/147 (51%), Positives = 107/147 (72%), Gaps = 2/147 (1%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
TID IL G+L EGD+I +CG +GPI+TTI+ LL PQPLKELRVK +YI V AS+G+
Sbjct 309 TIDAILSNGMLKEGDRIGVCGFNGPIVTTIKALLMPQPLKELRVKSQYIMVKEVRASLGI 368
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSL 121
KI A GL++A+AG+ + +VE D+ +++ + D+ +V S++ + GV V +STLGSL
Sbjct 369 KIAAGGLEKAIAGSRVLIVE--DNEEEVKRALETDLESVFSSIELSQEGVHVVSSTLGSL 426
Query 122 EALLVYLEECKIPVFAVSIGTVQKKDI 148
EAL+ +L+ KIP+ SIGTV+KKD+
Sbjct 427 EALISFLKTSKIPILGASIGTVKKKDM 453
> Hs15451892
Length=1220
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 75/156 (48%), Positives = 105/156 (67%), Gaps = 0/156 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL+ G L EGD I++ G+ GPI+T IR LL P P+KELRVK +Y H VEA+ G
Sbjct 871 TTIDVILINGRLKEGDTIIVPGVEGPIVTQIRGLLLPPPMKELRVKNQYEKHKEVEAAQG 930
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI+ L++ +AG L V + D I ++ E++ ++ L ++ GV+V ASTLGS
Sbjct 931 VKILGKDLEKTLAGLPLLVAYKEDEIPVLKDELIHELKQTLNAIKLEEKGVYVQASTLGS 990
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
LEALL +L+ ++P ++IG V KKD+ +ASVM E
Sbjct 991 LEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLE 1026
> 7292368
Length=932
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 64/134 (47%), Positives = 88/134 (65%), Gaps = 0/134 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTID IL+ G L EG +V+ G GPI+T IR+LL PQP+KELRVK Y+ + V+A+ G
Sbjct 795 TTIDAILINGKLREGQTMVVAGTDGPIVTQIRSLLMPQPMKELRVKNAYVEYKEVKAAQG 854
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI A L++A+AG +L + + D + EV +++ + L + +GV V ASTLGS
Sbjct 855 VKIAAKDLEKAIAGINLLIAHKPDEVEICTEEVARELKSALSHIKLAQTGVHVQASTLGS 914
Query 121 LEALLVYLEECKIP 134
LEALL +L KIP
Sbjct 915 LEALLEFLRTSKIP 928
> CE27011
Length=1173
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 57/156 (36%), Positives = 84/156 (53%), Gaps = 21/156 (13%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVILV G + GD IV+ G G I T +R LL P+PLKELRVK EYI + V+ + G
Sbjct 845 TTIDVILVNGTMRAGDVIVLTGSDGAITTQVRELLMPKPLKELRVKNEYIHYKEVKGARG 904
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VK++A L++ +AG +++ + D + + E + + L ++ G
Sbjct 905 VKVLAKNLEKVLAGLPIYITDREDEVDYLRHEADRQLANALHAIRKKPEG---------- 954
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
IP V+IG V KKD+++AS M+E
Sbjct 955 -----------NIPYSNVNIGPVHKKDVQKASAMKE 979
> Hs11345486
Length=538
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 81 EEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSLEA 123
EE + + GE K + AV + DS GSGV V L L A
Sbjct 122 EEPGPLKESPGEAFKALSAVEEECDSVGSGVQVVIEELRQLGA 164
> At3g56960
Length=779
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 11/45 (24%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 64 VAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTG 108
V+ G+D A +++ E G+ +DI+ +D+ A + +++ G
Sbjct 314 VSVGVDRAFEKMNMWGTESGEGAADIDSTTRRDLDAEMMRLEAEG 358
Lambda K H
0.317 0.136 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40