bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2476_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
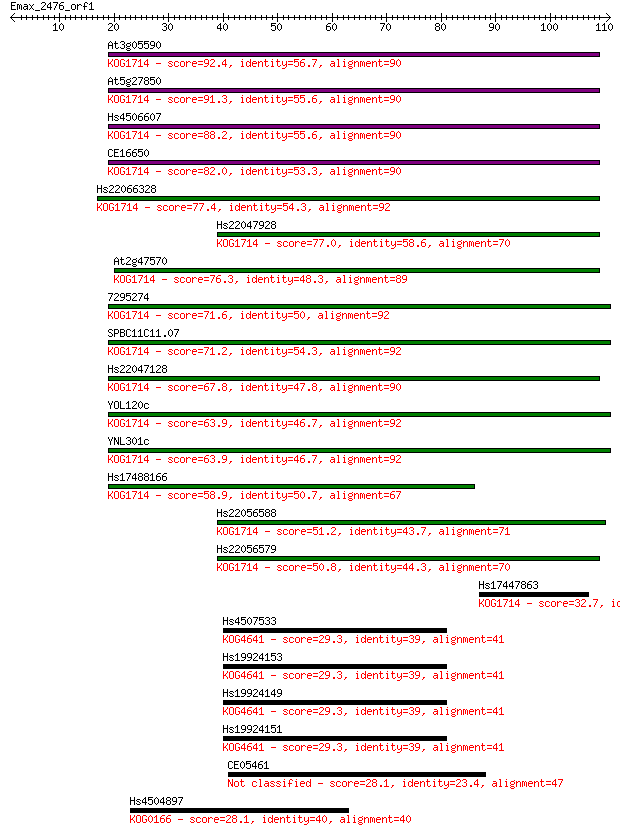
At3g05590 92.4 2e-19
At5g27850 91.3 4e-19
Hs4506607 88.2 3e-18
CE16650 82.0 3e-16
Hs22066328 77.4 6e-15
Hs22047928 77.0 8e-15
At2g47570 76.3 1e-14
7295274 71.6 4e-13
SPBC11C11.07 71.2 4e-13
Hs22047128 67.8 4e-12
YOL120c 63.9 6e-11
YNL301c 63.9 6e-11
Hs17488166 58.9 2e-09
Hs22056588 51.2 4e-07
Hs22056579 50.8 6e-07
Hs17447863 32.7 0.16
Hs4507533 29.3 1.8
Hs19924153 29.3 1.8
Hs19924149 29.3 1.8
Hs19924151 29.3 2.1
CE05461 28.1 3.7
Hs4504897 28.1 4.5
> At3g05590
Length=187
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 51/90 (56%), Positives = 64/90 (71%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL G++KK R A S++ YL+L VKLY+FL RRTNS FN VILKRL + +A
Sbjct 1 MGIDLIAGGKSKKTKRTAPKSDDVYLKLTVKLYRFLVRRTNSKFNGVILKRLFMSKVNKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLS+L M +E+K AV+VG+I DD R
Sbjct 61 PLSLSRLVEFMTGKEDKIAVLVGTITDDLR 90
> At5g27850
Length=187
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 50/90 (55%), Positives = 65/90 (72%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL G++KK R A S++ YL+L VKLY+FL RR+NSNFN VILKRL + +A
Sbjct 1 MGIDLIAGGKSKKTKRTAPKSDDVYLKLLVKLYRFLVRRSNSNFNAVILKRLFMSKVNKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLS+L M +++K AV+VG+I DD R
Sbjct 61 PLSLSRLVEFMTGKDDKIAVLVGTITDDLR 90
> Hs4506607
Length=188
Score = 88.2 bits (217), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 50/92 (54%), Positives = 67/92 (72%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG+D+++ + +K R+ S++ YLRL VKLY+FLARRTNS FN+V+LKRL R R
Sbjct 1 MGVDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSTFNQVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PLSLS++ M++ ENKTAVVVG+I DD R
Sbjct 60 PLSLSRMIRKMKLPGRENKTAVVVGTITDDVR 91
> CE16650
Length=188
Score = 82.0 bits (201), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 59/92 (64%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID+ N + R A SENPYLRL KLY FLARRT FN ++LKRL RR R
Sbjct 1 MGIDI-NHKHDRVARRTAPKSENPYLRLLSKLYAFLARRTGEKFNAIVLKRLRMSRRNRQ 59
Query 79 PLSLSKLSMRMRK--EENKTAVVVGSIVDDPR 108
PLSL+KL+ ++K ENKT V + ++ DD R
Sbjct 60 PLSLAKLARAVQKAGNENKTVVTLSTVTDDAR 91
> Hs22066328
Length=189
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 50/95 (52%), Positives = 63/95 (66%), Gaps = 5/95 (5%)
Query 17 AKMGID-LKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRR 75
A +G+D L N+ R K R+ S + YLRL VKLY+FLARRTNS FN+V+LKRL R
Sbjct 19 AIIGVDILHNKDR--KVPRKEPKSPDTYLRLLVKLYRFLARRTNSTFNQVVLKRLFRSRT 76
Query 76 LRAPLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PL L ++ M++ ENKTAVVVG+I DD R
Sbjct 77 KGPPLPLFRIIRKMKLPGRENKTAVVVGTIRDDVR 111
> Hs22047928
Length=210
Score = 77.0 bits (188), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 41/72 (56%), Positives = 53/72 (73%), Gaps = 2/72 (2%)
Query 39 SENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPLSLSKL--SMRMRKEENKT 96
S++ YLRL VKLY+FLAR TNS FN+V+LKRL R + PLSL ++ M++ ENKT
Sbjct 29 SQDIYLRLLVKLYRFLARLTNSTFNQVVLKRLFMSRTKQPPLSLCQMIRMMKLLGRENKT 88
Query 97 AVVVGSIVDDPR 108
AVVVG+ +DD R
Sbjct 89 AVVVGTTMDDVR 100
> At2g47570
Length=187
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 58/89 (65%), Gaps = 0/89 (0%)
Query 20 GIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAP 79
GIDL G++KK R S++ YL+L VK ++L RRT S FN VILKRL + +AP
Sbjct 1 GIDLIAGGKSKKTKRTEPKSDDVYLKLLVKANRYLVRRTESKFNAVILKRLFMSKVNKAP 60
Query 80 LSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
LSLS+L M ++ K AV+VG++ DD R
Sbjct 61 LSLSRLVRYMDGKDGKIAVIVGTVTDDVR 89
> 7295274
Length=188
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 60/95 (63%), Gaps = 5/95 (5%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID+ N +K R S++ YLRL VKLY+FL RRTN FN++ILKRL + R
Sbjct 1 MGIDI-NHKYDRKVRRTEPKSQDVYLRLLVKLYRFLQRRTNKKFNRIILKRLFMSKINRP 59
Query 79 PLSLSKLSMRMRKEENK---TAVVVGSIVDDPRTL 110
PLSL +++ R K N+ T VVVG++ DD R L
Sbjct 60 PLSLQRIA-RFFKAANQPESTIVVVGTVTDDARLL 93
> SPBC11C11.07
Length=187
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 50/96 (52%), Positives = 63/96 (65%), Gaps = 5/96 (5%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID++ R KK R SEN YL+L VKLY+FLARRT+S FNK ILKRL + R
Sbjct 1 MGIDIE-RHHVKKSQRSKPASENVYLKLLVKLYRFLARRTDSRFNKAILKRLFQSKTNRP 59
Query 79 PLSLSKL----SMRMRKEENKTAVVVGSIVDDPRTL 110
P+S+SK+ S + +NKT VVVG++ DD R L
Sbjct 60 PISISKIAALTSRKSASSQNKTTVVVGTVTDDERML 95
> Hs22047128
Length=143
Score = 67.8 bits (164), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 43/93 (46%), Positives = 61/93 (65%), Gaps = 5/93 (5%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAV-KLYQFLARRTNSNFNKVILKRLMAPRRLR 77
MG+D +++ R K R+ S++ YLRL+V + Q ++ TNS FN+V+LKRL R R
Sbjct 1 MGVDTRHKDR--KVRRKEPKSQDIYLRLSVGQAVQVSSKGTNSTFNRVVLKRLFMSRNNR 58
Query 78 APLSLSKLSMRMR--KEENKTAVVVGSIVDDPR 108
PL LS++ +M+ ENKTAVVVG+I DD R
Sbjct 59 PPLPLSRMIWKMKLPGRENKTAVVVGAITDDVR 91
> YOL120c
Length=186
Score = 63.9 bits (154), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/94 (45%), Positives = 62/94 (65%), Gaps = 2/94 (2%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID ++ + R A S+N YL+L VKLY FLARRT++ FNKV+LK L + R
Sbjct 1 MGIDHTSKQHKRSGHRTAPKSDNVYLKLLVKLYTFLARRTDAPFNKVVLKALFLSKINRP 60
Query 79 PLSLSKLSMRMRKE--ENKTAVVVGSIVDDPRTL 110
P+S+S+++ +++E NKT VVVG++ DD R
Sbjct 61 PVSVSRIARALKQEGAANKTVVVVGTVTDDARIF 94
> YNL301c
Length=186
Score = 63.9 bits (154), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/94 (45%), Positives = 62/94 (65%), Gaps = 2/94 (2%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID ++ + R A S+N YL+L VKLY FLARRT++ FNKV+LK L + R
Sbjct 1 MGIDHTSKQHKRSGHRTAPKSDNVYLKLLVKLYTFLARRTDAPFNKVVLKALFLSKINRP 60
Query 79 PLSLSKLSMRMRKE--ENKTAVVVGSIVDDPRTL 110
P+S+S+++ +++E NKT VVVG++ DD R
Sbjct 61 PVSVSRIARALKQEGAANKTVVVVGTVTDDARIF 94
> Hs17488166
Length=147
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 46/67 (68%), Gaps = 1/67 (1%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG D+ + + +K R+ S++ YLRL VKL FLARRT+S FN+V+LKRL R R
Sbjct 1 MGADICHN-KDRKVRRKEPKSQDIYLRLLVKLCSFLARRTDSTFNQVVLKRLFMSRTNRP 59
Query 79 PLSLSKL 85
PLSLS++
Sbjct 60 PLSLSQM 66
> Hs22056588
Length=153
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 40/71 (56%), Gaps = 18/71 (25%)
Query 39 SENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPLSLSKLSMRMRKEENKTAV 98
S + YLRL VKLY FLAR+ NS FN++I K M++ ENK AV
Sbjct 19 SRDIYLRLLVKLYSFLARQINSIFNQMIRK------------------MKLPGRENKMAV 60
Query 99 VVGSIVDDPRT 109
V+G+I+DD R
Sbjct 61 VLGTIMDDVRV 71
> Hs22056579
Length=209
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 40/70 (57%), Gaps = 18/70 (25%)
Query 39 SENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPLSLSKLSMRMRKEENKTAV 98
S + YLRL VKLY FLAR+ NS FN++I K M++ ENK AV
Sbjct 75 SRDIYLRLLVKLYSFLARQINSIFNQMIRK------------------MKLPGRENKMAV 116
Query 99 VVGSIVDDPR 108
V+G+I+DD R
Sbjct 117 VLGTIMDDVR 126
> Hs17447863
Length=119
Score = 32.7 bits (73), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 87 MRMRKEENKTAVVVGSIVDD 106
M++ +ENKTAVVVG+I DD
Sbjct 1 MKLPGQENKTAVVVGTITDD 20
> Hs4507533
Length=799
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 40 ENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPL 80
E LR V+LY+ L+R T + +L R + RRLR L
Sbjct 735 EKTLLRQQVELYRLLSRNTYLEWEDSVLGRHIFWRRLRKAL 775
> Hs19924153
Length=639
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 40 ENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPL 80
E LR V+LY+ L+R T + +L R + RRLR L
Sbjct 575 EKTLLRQQVELYRLLSRNTYLEWEDSVLGRHIFWRRLRKAL 615
> Hs19924149
Length=839
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 40 ENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPL 80
E LR V+LY+ L+R T + +L R + RRLR L
Sbjct 775 EKTLLRQQVELYRLLSRNTYLEWEDSVLGRHIFWRRLRKAL 815
> Hs19924151
Length=782
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 40 ENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPL 80
E LR V+LY+ L+R T + +L R + RRLR L
Sbjct 718 EKTLLRQQVELYRLLSRNTYLEWEDSVLGRHIFWRRLRKAL 758
> CE05461
Length=340
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 41 NPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPLSLSKLSM 87
+P+L ++L L+ + + +++++R + P + P SLS +S+
Sbjct 128 SPFLASLIRLVMILSPHNHGKYCRLLMRRFVFPFIIIVPFSLSLISL 174
> Hs4504897
Length=529
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 23 LKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
KN+G+ RR + N LR A K Q L RR S+F
Sbjct 17 FKNKGKDSTEMRRRRIEVNVELRKAKKDDQMLKRRNVSSF 56
Lambda K H
0.319 0.134 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40