bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2460_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
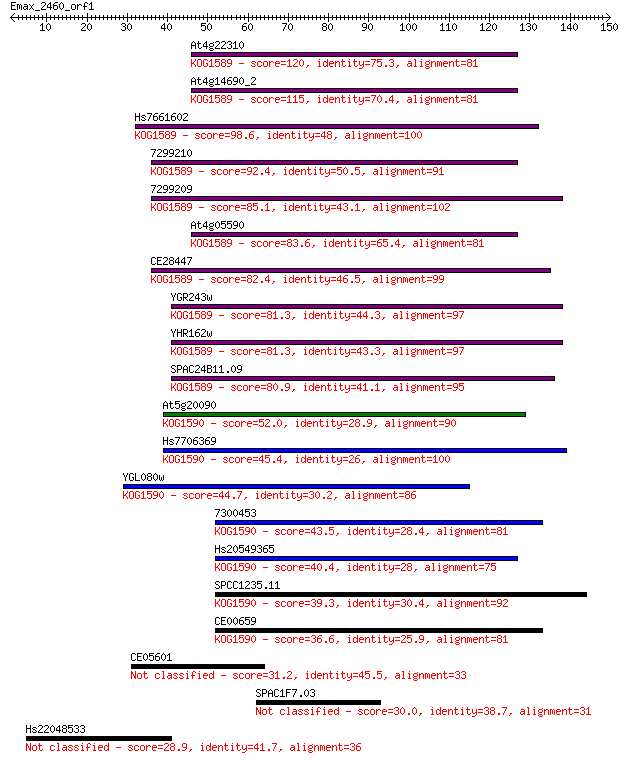
At4g22310 120 7e-28
At4g14690_2 115 2e-26
Hs7661602 98.6 4e-21
7299210 92.4 3e-19
7299209 85.1 5e-17
At4g05590 83.6 1e-16
CE28447 82.4 3e-16
YGR243w 81.3 6e-16
YHR162w 81.3 6e-16
SPAC24B11.09 80.9 9e-16
At5g20090 52.0 4e-07
Hs7706369 45.4 3e-05
YGL080w 44.7 6e-05
7300453 43.5 1e-04
Hs20549365 40.4 0.001
SPCC1235.11 39.3 0.003
CE00659 36.6 0.017
CE05601 31.2 0.67
SPAC1F7.03 30.0 1.6
Hs22048533 28.9 3.8
> At4g22310
Length=108
Score = 120 bits (302), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 61/81 (75%), Positives = 63/81 (77%), Gaps = 1/81 (1%)
Query 46 HPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPK 105
HPAGP TIHFWAPTFKWGISIANI D K +S QQ AV TG+IWSRYSMVI PK
Sbjct 12 HPAGPKTIHFWAPTFKWGISIANIADFA-KPPEKLSYPQQIAVTCTGVIWSRYSMVINPK 70
Query 106 NWNLFSVNVAMALTGCVQLAR 126
NWNLFSVNVAMA TG QLAR
Sbjct 71 NWNLFSVNVAMAGTGIYQLAR 91
> At4g14690_2
Length=122
Score = 115 bits (289), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 57/81 (70%), Positives = 60/81 (74%), Gaps = 1/81 (1%)
Query 46 HPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPK 105
HPAGP TIHFWAPTFKWGISIANI D K +S QQ AV TG+IW R S +ITPK
Sbjct 25 HPAGPKTIHFWAPTFKWGISIANIADF-QKPPENISYLQQIAVTCTGMIWCRCSTIITPK 83
Query 106 NWNLFSVNVAMALTGCVQLAR 126
NWNLFSVNVAMA TG QL R
Sbjct 84 NWNLFSVNVAMAATGIYQLTR 104
> Hs7661602
Length=127
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 48/100 (48%), Positives = 62/100 (62%), Gaps = 1/100 (1%)
Query 32 LQLPLPERAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAAT 91
++L LPE+ + HPAGP T+ FWAP KWG+ A + DM + +S AQ + AT
Sbjct 20 VELMLPEKLRPLYNHPAGPRTVFFWAPIMKWGLVCAGLADM-ARPAEKLSTAQSAVLMAT 78
Query 92 GIIWSRYSMVITPKNWNLFSVNVAMALTGCVQLARIALKN 131
G IWSRYS+VI PKNW+LF+VN + G QL RI N
Sbjct 79 GFIWSRYSLVIIPKNWSLFAVNFFVGAAGASQLFRIWRYN 118
> 7299210
Length=154
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 46/91 (50%), Positives = 63/91 (69%), Gaps = 1/91 (1%)
Query 36 LPERAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIW 95
+P + + HPAGP TI FWAP FKWG+ A + D+ + +S + A+AATGIIW
Sbjct 43 VPAKLRPLWMHPAGPKTIFFWAPVFKWGLVAAGLSDL-ARPADTISVSGCAALAATGIIW 101
Query 96 SRYSMVITPKNWNLFSVNVAMALTGCVQLAR 126
SRYS+VI PKN++LF+VN+ + +T VQLAR
Sbjct 102 SRYSLVIIPKNYSLFAVNLFVGITQVVQLAR 132
> 7299209
Length=151
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 63/102 (61%), Gaps = 3/102 (2%)
Query 36 LPERAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIW 95
+P + + HPAGP TI FWAP KW + IA + D+ + +SP A+ AT +IW
Sbjct 37 VPPKMRPLWMHPAGPKTIFFWAPIVKWSLVIAGLSDL-TRPADKISPNGCLALGATNLIW 95
Query 96 SRYSMVITPKNWNLFSVNVAMALTGCVQLARIALKNFQNSSS 137
+RYS+VI PKN++LF+VN+ ++LT QL R N+Q S
Sbjct 96 TRYSLVIIPKNYSLFAVNLFVSLTQLFQLGRYY--NYQWEQS 135
> At4g05590
Length=150
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 53/137 (38%), Positives = 62/137 (45%), Gaps = 57/137 (41%)
Query 46 HPAGPFT------------------IHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQ-- 85
HPAGP T +HFWAPTFKWGISIANI D K +S QQ
Sbjct 12 HPAGPKTSESSISMNVFFFTFDFSHVHFWAPTFKWGISIANIADF-QKPPETLSYPQQIG 70
Query 86 ------------------------------------TAVAATGIIWSRYSMVITPKNWNL 109
+ + TG++WSRYS VITPKNWNL
Sbjct 71 IILTIGLSYLYSAHIAVMYRCVNFNVYMMPRMAVIVSVITGTGLVWSRYSTVITPKNWNL 130
Query 110 FSVNVAMALTGCVQLAR 126
FSV++ MA+TG QL R
Sbjct 131 FSVSLGMAVTGIYQLTR 147
> CE28447
Length=133
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 46/99 (46%), Positives = 59/99 (59%), Gaps = 1/99 (1%)
Query 36 LPERAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIW 95
LP AK H AGP T+ FWAPT KW + A + D+ + +S Q +A+ ATG IW
Sbjct 23 LPAFAKPAWNHAAGPKTVFFWAPTIKWTLIGAGLADLA-RPADKLSLYQNSALFATGAIW 81
Query 96 SRYSMVITPKNWNLFSVNVAMALTGCVQLARIALKNFQN 134
+RY +VITP N+ L SVN + TG QL RIA +QN
Sbjct 82 TRYCLVITPINYYLSSVNFFVMCTGLAQLCRIAHYRYQN 120
> YGR243w
Length=146
Score = 81.3 bits (199), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 43/97 (44%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 41 KNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSM 100
+ F GP T+HFWAPT KWG+ A + D I + V VS AQ ++ AT +IW+R+S
Sbjct 11 RRFWNSETGPKTVHFWAPTLKWGLVFAGLND-IKRPVEKVSGAQNLSLLATALIWTRWSF 69
Query 101 VITPKNWNLFSVNVAMALTGCVQLARIALKNFQNSSS 137
VI PKN+ L SVN + T L RIA +N S
Sbjct 70 VIKPKNYLLASVNFFLGCTAGYHLTRIANFRIRNGDS 106
> YHR162w
Length=129
Score = 81.3 bits (199), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 42/97 (43%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 41 KNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSM 100
+ F GP T+HFWAPT KWG+ A DM + V +S AQ ++ +T +IW+R+S
Sbjct 11 RRFWQSETGPKTVHFWAPTLKWGLVFAGFSDM-KRPVEKISGAQNLSLLSTALIWTRWSF 69
Query 101 VITPKNWNLFSVNVAMALTGCVQLARIALKNFQNSSS 137
VI P+N L SVN + LT QL RIA +N S
Sbjct 70 VIKPRNILLASVNSFLCLTAGYQLGRIANYRIRNGDS 106
> SPAC24B11.09
Length=118
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 39/95 (41%), Positives = 58/95 (61%), Gaps = 1/95 (1%)
Query 41 KNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSM 100
K F HPAGP T+HFWAP KW + ++ I D + +S Q A+ ATG IW+R+S+
Sbjct 7 KRFWNHPAGPKTVHFWAPAMKWTLVLSGIGDY-ARSPEYLSIRQYAALCATGAIWTRWSL 65
Query 101 VITPKNWNLFSVNVAMALTGCVQLARIALKNFQNS 135
++ PKN+ +VN +A+ G VQ++RI + Q
Sbjct 66 IVRPKNYFNATVNFFLAIVGAVQVSRILVYQRQQK 100
> At5g20090
Length=110
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 45/90 (50%), Gaps = 1/90 (1%)
Query 39 RAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRY 98
R + F+ P GP T HFW P WG A ++DM K +S +A+ ++ R+
Sbjct 5 RFQAFLNSPIGPKTTHFWGPIANWGFVAAGLVDM-QKPPEMISGNMSSAMCVYSALFMRF 63
Query 99 SMVITPKNWNLFSVNVAMALTGCVQLARIA 128
+ ++ P+N+ L + + + QL+R A
Sbjct 64 AWMVQPRNYLLLACHASNETVQLYQLSRWA 93
> Hs7706369
Length=109
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 47/100 (47%), Gaps = 4/100 (4%)
Query 39 RAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRY 98
R+K+F + + HFW P WG+ IA I DM K +S A+ + + R+
Sbjct 14 RSKDFRDYL---MSTHFWGPVANWGLPIAAINDM-KKSPEIISGRMTFALCCYSLTFMRF 69
Query 99 SMVITPKNWNLFSVNVAMALTGCVQLARIALKNFQNSSSS 138
+ + P+NW LF+ + + +Q R+ ++S+
Sbjct 70 AYKVQPRNWLLFACHATNEVAQLIQGGRLIKHEMTKTASA 109
> YGL080w
Length=130
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 44/86 (51%), Gaps = 7/86 (8%)
Query 29 RRLLQLPLPERAKNFIGHPAGPFTIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAV 88
R LQ + + +I FT HFW P +GI IA I D+ KD + +S A+
Sbjct 11 RSFLQKYINKETLKYI------FTTHFWGPVSNFGIPIAAIYDL-KKDPTLISGPMTFAL 63
Query 89 AATGIIWSRYSMVITPKNWNLFSVNV 114
++ +Y++ ++PKN+ LF ++
Sbjct 64 VTYSGVFMKYALSVSPKNYLLFGCHL 89
> 7300453
Length=107
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 1/81 (1%)
Query 52 TIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPKNWNLFS 111
+ HFW P WGI +A + D K +S A+ I+ R++ + P+NW LF+
Sbjct 21 STHFWGPVANWGIPVAALAD-TQKSPKFISGKMTLALTLYSCIFMRFAYKVQPRNWLLFA 79
Query 112 VNVAMALTGCVQLARIALKNF 132
+ A +Q R N+
Sbjct 80 CHATNATAQSIQGLRFLHYNY 100
> Hs20549365
Length=323
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query 52 TIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPKNWNLFS 111
+ HFW P F WG+ +A DM +S TA+ I+ R++ + P+N L +
Sbjct 213 STHFWGPAFSWGLPLAAFKDM-KASPEIISGRMTTALILYSAIFMRFAYRVQPRNLLLMA 271
Query 112 VNVAMALTGCVQLAR 126
+ + VQ +R
Sbjct 272 CHCTNVMAQSVQASR 286
> SPCC1235.11
Length=141
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 44/93 (47%), Gaps = 5/93 (5%)
Query 52 TIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPKNWNLFS 111
+ HFW P +GI IA I+D+ KD +S A+ ++ RY+ +++P+N+ L
Sbjct 34 STHFWGPLSNFGIPIAAILDL-KKDPRLISGRMTGALILYSSVFMRYAWMVSPRNYLLLG 92
Query 112 VNVAMALTGCVQLAR-IALKNFQNSSSSGSNSS 143
+ A VQ A+ I NF S S
Sbjct 93 CH---AFNTTVQTAQGIRFVNFWYGKEGASKQS 122
> CE00659
Length=160
Score = 36.6 bits (83), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 39/81 (48%), Gaps = 1/81 (1%)
Query 52 TIHFWAPTFKWGISIANIIDMIDKDVSGVSPAQQTAVAATGIIWSRYSMVITPKNWNLFS 111
+ HFW P WG+ +A + D + K+ +S +A+ ++ R++ + P+N LF+
Sbjct 26 STHFWGPVANWGLPLAALGD-LKKNPDMISGPMTSALLIYSSVFMRFAWHVQPRNLLLFA 84
Query 112 VNVAMALTGCVQLARIALKNF 132
+ A QL R N+
Sbjct 85 CHFANFSAQGAQLGRFVNHNY 105
> CE05601
Length=675
Score = 31.2 bits (69), Expect = 0.67, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query 31 LLQLPLPERAKNFIGHPAGPFTIHF-WAPTFKWG 63
++ L +AKNF+ HPA I F W P FK G
Sbjct 129 VMALASAAKAKNFLSHPACQREIDFRWQPGFKLG 162
> SPAC1F7.03
Length=710
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 62 WGISIANIIDMIDKDVSGVSPAQQTAVAATG 92
+GI I +ID D ++SG P Q +A +G
Sbjct 81 YGIEIKKVIDPCDMEISGFCPMQTGNIALSG 111
> Hs22048533
Length=191
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 5 AAAAAAAAATGRSLFFPGIVPPLQRRLLQLPLPERA 40
A A A TGR L+ +VPP Q ++L + PE A
Sbjct 132 GAEHAGAEVTGRRLYRMPVVPPTQPKMLFIQSPEPA 167
Lambda K H
0.320 0.129 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40