bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2453_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
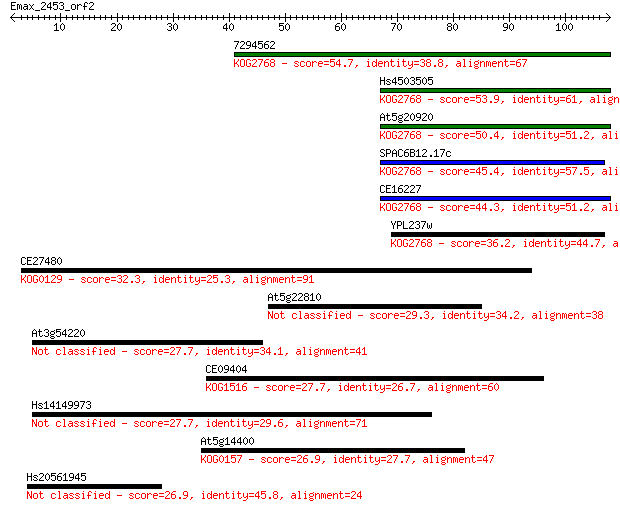
7294562 54.7 4e-08
Hs4503505 53.9 7e-08
At5g20920 50.4 7e-07
SPAC6B12.17c 45.4 2e-05
CE16227 44.3 5e-05
YPL237w 36.2 0.013
CE27480 32.3 0.20
At5g22810 29.3 2.0
At3g54220 27.7 4.9
CE09404 27.7 5.1
Hs14149973 27.7 5.9
At5g14400 26.9 8.7
Hs20561945 26.9 9.6
> 7294562
Length=312
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 42/69 (60%), Gaps = 2/69 (2%)
Query 41 QEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIVKNNPDLAGSK--RYTIKPP 98
Q D ++ E D S F YTY+E+L R+ ++I+ NPD+A + ++ ++PP
Sbjct 127 QADDDKSEDKENDEDNSSTWFGSDRDYTYDELLKRVFEIILDKNPDMAAGRKPKFVMRPP 186
Query 99 QVVRVGSKK 107
QV+RVG+KK
Sbjct 187 QVLRVGTKK 195
> Hs4503505
Length=333
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTYEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 174 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 216
> At5g20920
Length=268
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 34/42 (80%), Gaps = 1/42 (2%)
Query 67 YTYEEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVRVGSKK 107
Y Y+E+L R+ +++ +NNP+LAG +R T+ +PPQV+R G+KK
Sbjct 116 YIYDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKK 157
> SPAC6B12.17c
Length=310
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/41 (56%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 67 YTYEEMLSRIQDLIVKNNPDLAGSKR-YTIKPPQVVRVGSK 106
Y Y E+L+R L+ NNP+LAG KR YTI PP V R G K
Sbjct 157 YYYPELLNRFFTLLRTNNPELAGEKRKYTIVPPSVHREGKK 197
> CE16227
Length=250
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 67 YTYEEMLSRIQDLIVKNNPDLAGS-KRYTIKPPQVVRVGSKK 107
YTYEE L+ + ++ NPD AG K++ IK P+V R GSKK
Sbjct 87 YTYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKK 128
> YPL237w
Length=285
Score = 36.2 bits (82), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 3/41 (7%)
Query 69 YEEMLSRIQDLIVKNNPDLAGSK---RYTIKPPQVVRVGSK 106
Y E+LSR +++ NNP+LAG + ++ I PP +R G K
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDGKK 171
> CE27480
Length=619
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query 3 CCSPPPPAPDTAATAEAQQPAAQAFDFGERRKKKKEKKQ---EDQGEAAAAEGFIDGSGQ 59
CC+P PP + Q PAA DFG + + E + G+
Sbjct 121 CCNPAPPY-----ASHCQSPAASD-DFGHHFGSFDSMSAYNFHNNNQQQRREAHVSRDGK 174
Query 60 IFVRGNVYTYEEMLSRIQDLIVKNNPDLAGSKRY 93
F +GN+YT E++ + +++ +NN + +K +
Sbjct 175 SFDQGNIYTAEDVHTVVKNHRNENNQRVVSNKVF 208
> At5g22810
Length=337
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 26/42 (61%), Gaps = 4/42 (9%)
Query 47 AAAAEGFIDGSGQIF----VRGNVYTYEEMLSRIQDLIVKNN 84
A+AA G+ DG+ +++ + + Y++ +SRIQ++ NN
Sbjct 93 ASAASGYYDGTAKLYSAISLPQQLEHYKDYISRIQEIATSNN 134
> At3g54220
Length=653
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query 5 SPPPPAPDTAATAEAQQPAAQAFDFGERRKKKKEKKQEDQG 45
+PP P TA Q A+A ER+++ K +KQ+++G
Sbjct 249 APPQPETVTATVPAVQTNTAEALR--ERKEEIKRQKQDEEG 287
> CE09404
Length=583
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 36 KKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIVKNNPDLAGSKRYTI 95
KK ED E AA G+I +FV Y Y E ++++ + + + D + + I
Sbjct 414 KKLSDDEDDKEVAA-RGYIQLYSDLFVNNGTYNYAEKMTKLGNKVFMYSFDYCNPRSFGI 472
> Hs14149973
Length=178
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 31/72 (43%), Gaps = 1/72 (1%)
Query 5 SPPPPAPDTAATAE-AQQPAAQAFDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVR 63
SP P A DT TA QP+ G R+ K Q QGE+ E + S +
Sbjct 86 SPHPSASDTVGTAGLGVQPSRHWSVSGGPRQPKSSGSQGPQGESLDKEAWALRSSTVSAG 145
Query 64 GNVYTYEEMLSR 75
++++E + R
Sbjct 146 ARRWSWDECVDR 157
> At5g14400
Length=432
Score = 26.9 bits (58), Expect = 8.7, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 35 KKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIV 81
K++E+++ED A E F+D + YEE +S + D+++
Sbjct 204 KEREREEEDMNNAIREEDFLDS----IISNEDLNYEEKVSIVLDILL 246
> Hs20561945
Length=1260
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 4 CSPPPPAPDTAATAEAQQPAAQAF 27
CSP P AP+ Q+PA QA+
Sbjct 104 CSPKPQAPEQRRACPPQRPAPQAW 127
Lambda K H
0.312 0.130 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40