bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2372_orf1
Length=60
Score E
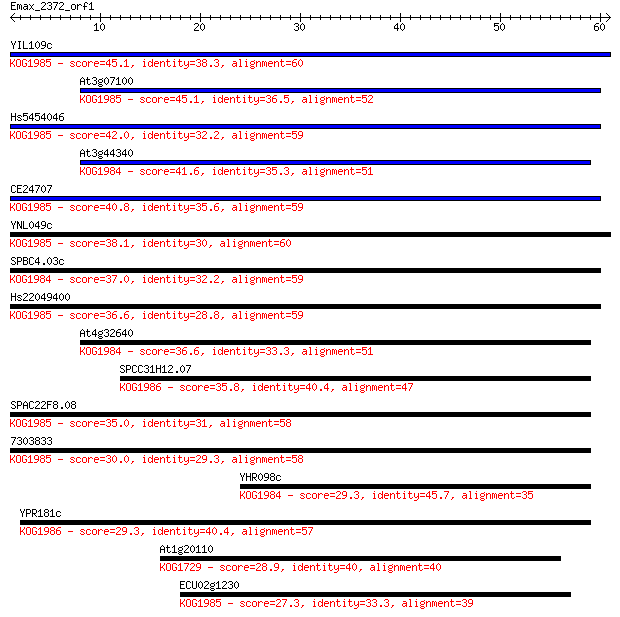
Sequences producing significant alignments: (Bits) Value
YIL109c 45.1 3e-05
At3g07100 45.1 3e-05
Hs5454046 42.0 3e-04
At3g44340 41.6 3e-04
CE24707 40.8 7e-04
YNL049c 38.1 0.004
SPBC4.03c 37.0 0.009
Hs22049400 36.6 0.010
At4g32640 36.6 0.012
SPCC31H12.07 35.8 0.021
SPAC22F8.08 35.0 0.034
7303833 30.0 1.1
YHR098c 29.3 1.7
YPR181c 29.3 1.8
At1g20110 28.9 2.4
ECU02g1230 27.3 7.0
> YIL109c
Length=926
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 37/60 (61%), Gaps = 0/60 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLRR 60
++ I L IP I++SN T+ LG A++SA + VGGKI++ + + P +G G L+R
Sbjct 400 RQNIETLLTKIPQIFQSNLITNFALGPALKSAYHLIGGVGGKIIVVSGTLPNLGIGKLQR 459
> At3g07100
Length=1054
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 36/52 (69%), Gaps = 0/52 (0%)
Query 8 LNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLR 59
L+++P +++ N ++ G A+R+A MV +GGK+L+F +S P++G G L+
Sbjct 539 LDSLPLMFQDNFNVESAFGPALRAAFMVMNQLGGKLLIFQNSLPSLGAGRLK 590
> Hs5454046
Length=1268
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLR 59
KE I + LNA+P+++ + T + LG A+++A + GG++ +F + P++G G+L+
Sbjct 766 KELIKDLLNALPNMFTNTRETHSALGPALQAAFKLMSPTGGRVSVFQTQLPSLGAGLLQ 824
> At3g44340
Length=1122
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 8 LNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
L +IP +++ + ++ G+AV++A + K GGK+++F S P+VG G L
Sbjct 614 LESIPTMFQESKSPESAFGAAVKAAFLAMKSTGGKLMVFQSVLPSVGIGAL 664
> CE24707
Length=1024
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query 1 KEAILNTLNAIPDIWKSNACTDN--CLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
K+ I + +P+ + T N CLGSA++ A + + VGG+I +F S P +G G L
Sbjct 494 KDTIRTFIKQLPEFYSQIGPTSNGNCLGSALKLAQSMIQEVGGRISIFQVSLPNLGLGAL 553
Query 59 R 59
+
Sbjct 554 K 554
> YNL049c
Length=876
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLRR 60
K + L IP+I++ + LG A+++A+ + K GGK+ + +S+ P G G L++
Sbjct 356 KNNLETLLKKIPEIFQDTHSSKFALGPALKAASNLIKSTGGKVEVISSTLPNTGIGKLKK 415
> SPBC4.03c
Length=891
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLR 59
K I L++ P I++ + +G+A+ ++ K GGK +F S+ PT+G G LR
Sbjct 381 KVVIERLLDSFPAIFQDVKIPEPVVGNALEVVKLLLKETGGKAFVFVSALPTMGLGKLR 439
> Hs22049400
Length=1093
Score = 36.6 bits (83), Expect = 0.010, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVLR 59
KE + + L +P ++ T + LG A+++A + GG++ +F + PT+G G L+
Sbjct 592 KELVQDLLKTLPQMFTKTLETQSALGPALQAAFKLMSPTGGRMSVFQTQLPTLGVGALK 650
> At4g32640
Length=1069
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 8 LNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
L++IP +++ + ++ G+AV++A + K GGK+++F S +VG G L
Sbjct 600 LDSIPTMFQESKIPESAFGAAVKAAFLAMKSKGGKLMVFQSILCSVGVGAL 650
> SPCC31H12.07
Length=759
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 24/53 (45%), Gaps = 6/53 (11%)
Query 12 PDIW------KSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
PD W + CT L +V V + GG I+LFA P TVG G +
Sbjct 245 PDSWPVANDRRPQRCTGTALNISVSMMESVCPNSGGHIMLFAGGPSTVGPGTV 297
> SPAC22F8.08
Length=926
Score = 35.0 bits (79), Expect = 0.034, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
++ I N L ++ + + N LG +++A + +++GGKI SS P VG G L
Sbjct 419 RQGIENLLERFQSMFATTRDSSNALGPGLKAAHRLIENIGGKICCLISSLPNVGVGKL 476
> 7303833
Length=1184
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 1 KEAILNTLNAIPDIWKSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEGVL 58
KE + + L +P + + LG+A++ A + + GG+I +F S P G G L
Sbjct 673 KELVRDLLKQLPKRFAHTHDPGSALGAALQVAFKLMQASGGRITVFQSCLPNKGPGAL 730
> YHR098c
Length=929
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query 24 CLGSAVRSAAMVAKHV----GGKILLFASSPPTVGEGVL 58
C GSA+++A + V GGKI+ +S PT+G G L
Sbjct 412 CYGSALQAAKLALDTVTGGQGGKIICSLNSLPTIGNGNL 450
> YPR181c
Length=768
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Query 2 EAILNTL--NAIPDIWKSNACTD--NCLGSAVRSAAMVA----KHVGGKILLFASSPPTV 53
E LN L N PD W A GSA+ A+++ K++ +I+LFAS P TV
Sbjct 235 EFKLNQLLENLSPDQWSVPAGHRPLRATGSALNIASLLLQGCYKNIPARIILFASGPGTV 294
Query 54 GEGVL 58
G++
Sbjct 295 APGLI 299
> At1g20110
Length=613
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 5/40 (12%)
Query 16 KSNACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGE 55
+SN+ T N L V +A AK +GG +S PPT G+
Sbjct 389 QSNSYTTNTLLDTVTAAMFQAKEIGG-----SSRPPTSGK 423
> ECU02g1230
Length=705
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 18 NACTDNCLGSAVRSAAMVAKHVGGKILLFASSPPTVGEG 56
N T N G A+R + GG I+ F ++ P +G G
Sbjct 258 NKSTKNNYGDALRVVQSILGATGGCIISFLATHPNMGSG 296
Lambda K H
0.318 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181901402
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40