bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2302_orf1
Length=78
Score E
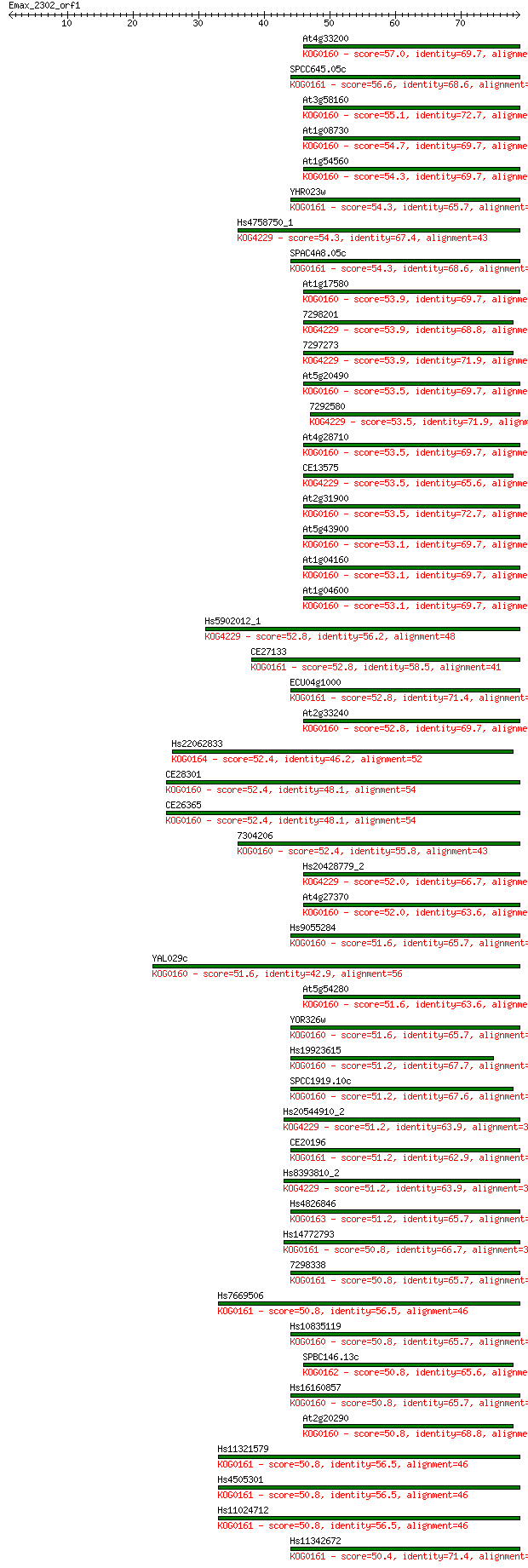
Sequences producing significant alignments: (Bits) Value
At4g33200 57.0 7e-09
SPCC645.05c 56.6 1e-08
At3g58160 55.1 3e-08
At1g08730 54.7 4e-08
At1g54560 54.3 5e-08
YHR023w 54.3 6e-08
Hs4758750_1 54.3 6e-08
SPAC4A8.05c 54.3 6e-08
At1g17580 53.9 6e-08
7298201 53.9 7e-08
7297273 53.9 8e-08
At5g20490 53.5 8e-08
7292580 53.5 9e-08
At4g28710 53.5 9e-08
CE13575 53.5 9e-08
At2g31900 53.5 1e-07
At5g43900 53.1 1e-07
At1g04160 53.1 1e-07
At1g04600 53.1 1e-07
Hs5902012_1 52.8 1e-07
CE27133 52.8 2e-07
ECU04g1000 52.8 2e-07
At2g33240 52.8 2e-07
Hs22062833 52.4 2e-07
CE28301 52.4 2e-07
CE26365 52.4 2e-07
7304206 52.4 2e-07
Hs20428779_2 52.0 3e-07
At4g27370 52.0 3e-07
Hs9055284 51.6 3e-07
YAL029c 51.6 3e-07
At5g54280 51.6 3e-07
YOR326w 51.6 4e-07
Hs19923615 51.2 4e-07
SPCC1919.10c 51.2 4e-07
Hs20544910_2 51.2 4e-07
CE20196 51.2 4e-07
Hs8393810_2 51.2 5e-07
Hs4826846 51.2 5e-07
Hs14772793 50.8 5e-07
7298338 50.8 5e-07
Hs7669506 50.8 6e-07
Hs10835119 50.8 6e-07
SPBC146.13c 50.8 6e-07
Hs16160857 50.8 6e-07
At2g20290 50.8 6e-07
Hs11321579 50.8 6e-07
Hs4505301 50.8 7e-07
Hs11024712 50.8 7e-07
Hs11342672 50.4 7e-07
> At4g33200
Length=1374
Score = 57.0 bits (136), Expect = 7e-09, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFECFK NS EQ CIN+ NE LQ++FN
Sbjct 428 GVLDIYGFECFKNNSFEQFCINFANEKLQQHFN 460
> SPCC645.05c
Length=1526
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 24/35 (68%), Positives = 30/35 (85%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F G+L+I GFE F++NS EQLCINYTNE LQ++FN
Sbjct 440 FIGILDIAGFEIFEKNSFEQLCINYTNEKLQQFFN 474
> At3g58160
Length=1242
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CINYTNE LQ++FN
Sbjct 429 GVLDIYGFESFKTNSFEQFCINYTNEKLQQHFN 461
> At1g08730
Length=1572
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 439 GVLDIYGFESFKTNSFEQFCINFTNEKLQQHFN 471
> At1g54560
Length=1529
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 434 GVLDIYGFESFKTNSFEQFCINFTNEKLQQHFN 466
> YHR023w
Length=1928
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 29/35 (82%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ G+L+I GFE F+ NS EQLCINYTNE LQ++FN
Sbjct 459 YIGLLDIAGFEIFENNSFEQLCINYTNEKLQQFFN 493
> Hs4758750_1
Length=1144
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 29/47 (61%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 36 NKKEEE----ILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
NKK+ E L GVL+IFGFE F+ NS EQ CINY NE LQ YFN
Sbjct 503 NKKDVEEAVSCLSIGVLDIFGFEDFERNSFEQFCINYANEQLQYYFN 549
> SPAC4A8.05c
Length=2104
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/35 (68%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F NS EQLCIN+TNE LQ++FN
Sbjct 453 FIGVLDIAGFEIFTFNSFEQLCINFTNEKLQQFFN 487
> At1g17580
Length=1536
Score = 53.9 bits (128), Expect = 6e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 443 GVLDIYGFESFKCNSFEQFCINFTNEKLQQHFN 475
> 7298201
Length=2167
Score = 53.9 bits (128), Expect = 7e-08, Method: Composition-based stats.
Identities = 22/32 (68%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
GVL+IFGFE F +NS EQ CINY NE LQ++F
Sbjct 428 GVLDIFGFENFDQNSFEQFCINYANENLQQFF 459
> 7297273
Length=2129
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 23/32 (71%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
GVL+IFGFE F NS EQLCINY NE LQ++F
Sbjct 439 GVLDIFGFENFDNNSFEQLCINYANENLQQFF 470
> At5g20490
Length=1544
Score = 53.5 bits (127), Expect = 8e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 450 GVLDIYGFESFKINSFEQFCINFTNEKLQQHFN 482
> 7292580
Length=2424
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/32 (71%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 47 VLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+L+IFGFE ENS EQLCINY NE LQ YFN
Sbjct 520 ILDIFGFEDLAENSFEQLCINYANENLQLYFN 551
> At4g28710
Length=899
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 430 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 462
> CE13575
Length=2098
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 21/32 (65%), Positives = 27/32 (84%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
G+L+IFGFE F+ NS EQLCIN+ NE LQ++F
Sbjct 427 GILDIFGFENFESNSFEQLCINFANETLQQFF 458
> At2g31900
Length=1490
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQLCIN TNE LQ++FN
Sbjct 365 GVLDIYGFESFKINSFEQLCINLTNEKLQQHFN 397
> At5g43900
Length=1505
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 431 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 463
> At1g04160
Length=1519
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 432 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 464
> At1g04600
Length=1730
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 430 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 462
> Hs5902012_1
Length=1325
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/48 (56%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Query 31 KEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
K+ E N K I GVL+IFGFE ++ NS EQ CIN+ NE LQ YFN
Sbjct 522 KDLEHNTKTLSI---GVLDIFGFEDYENNSFEQFCINFANERLQHYFN 566
> CE27133
Length=2003
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 38 KEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+++ + F G+L+I GFE F+ NS EQLCINYTNE LQ+ FN
Sbjct 461 RQQSVSFIGILDIAGFEIFETNSFEQLCINYTNEKLQQLFN 501
> ECU04g1000
Length=1700
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/35 (71%), Positives = 30/35 (85%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F++NS EQLCINYTNE LQ++FN
Sbjct 463 FIGVLDIAGFEIFEKNSFEQLCINYTNEKLQQFFN 497
> At2g33240
Length=1611
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 445 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 477
> Hs22062833
Length=930
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/52 (46%), Positives = 35/52 (67%), Gaps = 3/52 (5%)
Query 26 VKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
++ +G++ ++ K+ I GVL+I+GFE F NS EQ CINY NE LQ+ F
Sbjct 287 MEPRGRDPRRDGKDTVI---GVLDIYGFEVFPVNSFEQFCINYCNEKLQQLF 335
> CE28301
Length=1835
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 25 GVKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ +K K + N+K+ F GVL+I+GFE F NS EQ INY NE LQ+ FN
Sbjct 465 ALNEKDKLDGTNQKKRPDRFIGVLDIYGFETFDVNSFEQFSINYANEKLQQQFN 518
> CE26365
Length=1833
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 25 GVKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ +K K + N+K+ F GVL+I+GFE F NS EQ INY NE LQ+ FN
Sbjct 465 ALNEKDKLDGTNQKKRPDRFIGVLDIYGFETFDVNSFEQFSINYANEKLQQQFN 518
> 7304206
Length=1786
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 36 NKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
N ++ F GVL+I+GFE F+ NS EQ CINY NE LQ+ FN
Sbjct 434 NNGSKQCSFIGVLDIYGFETFEVNSFEQFCINYANEKLQQQFN 476
> Hs20428779_2
Length=921
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 22/33 (66%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFE F+ NS EQLCIN NE +Q YFN
Sbjct 362 GILDIFGFENFQRNSFEQLCINIANEQIQYYFN 394
> At4g27370
Length=1126
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/33 (63%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+L+I+GFE FK+NS EQ CINY NE LQ++FN
Sbjct 517 SILDIYGFESFKDNSFEQFCINYANERLQQHFN 549
> Hs9055284
Length=1742
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F NS EQ CINY NE LQ+ FN
Sbjct 430 FIGVLDIYGFETFDVNSFEQFCINYANEKLQQQFN 464
> YAL029c
Length=1471
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 23 LGGVKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ + K + E ++++ F G+L+I+GFE F++NS EQ CINY NE LQ+ FN
Sbjct 423 VDNINKTLYDPELDQQDHVFSFIGILDIYGFEHFEKNSFEQFCINYANEKLQQEFN 478
> At5g54280
Length=1111
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/33 (63%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+L+I+GFE FK NS EQ CINY NE LQ++FN
Sbjct 465 SILDIYGFESFKNNSFEQFCINYANERLQQHFN 497
> YOR326w
Length=1574
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F++NS EQ CINY NE LQ+ FN
Sbjct 443 FIGVLDIYGFEHFEKNSFEQFCINYANEKLQQEFN 477
> Hs19923615
Length=770
Score = 51.2 bits (121), Expect = 4e-07, Method: Composition-based stats.
Identities = 21/31 (67%), Positives = 26/31 (83%), Gaps = 0/31 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQ 74
F G+L+++GFE F +NSLEQLCINY NE LQ
Sbjct 409 FIGLLDVYGFESFPDNSLEQLCINYANEKLQ 439
> SPCC1919.10c
Length=1516
Score = 51.2 bits (121), Expect = 4e-07, Method: Composition-based stats.
Identities = 23/34 (67%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
F GVL+I+GFE FK+NS EQ CINY NE LQ+ F
Sbjct 452 FIGVLDIYGFEHFKKNSFEQFCINYANEKLQQEF 485
> Hs20544910_2
Length=1280
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 43 LFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
L G+L+IFGFE FK+NS EQLCIN NE +Q Y+N
Sbjct 375 LSIGILDIFGFENFKKNSFEQLCINIANEQIQYYYN 410
> CE20196
Length=1413
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/35 (62%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F VL+I GFE ++NS EQ CINYTNE LQ++FN
Sbjct 393 FIAVLDIAGFEIIEKNSFEQFCINYTNEKLQQFFN 427
> Hs8393810_2
Length=1279
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 43 LFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
L G+L+IFGFE FK+NS EQLCIN NE +Q Y+N
Sbjct 375 LSIGILDIFGFENFKKNSFEQLCINIANEQIQYYYN 410
> Hs4826846
Length=1285
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F+ NS EQ CINY NE LQ++FN
Sbjct 451 FIGVLDIAGFEYFEHNSFEQFCINYCNEKLQQFFN 485
> Hs14772793
Length=1220
Score = 50.8 bits (120), Expect = 5e-07, Method: Composition-based stats.
Identities = 24/36 (66%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 43 LFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F+ NS EQLCIN+TNE LQ++FN
Sbjct 156 FFIGVLDIAGFEIFEFNSFEQLCINFTNEKLQQFFN 191
> 7298338
Length=538
Score = 50.8 bits (120), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F+ N EQLCIN+TNE LQ++FN
Sbjct 455 FIGVLDIAGFEIFEYNGFEQLCINFTNEKLQQFFN 489
> Hs7669506
Length=1939
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 33 EEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
++ + K+ F GVL+I GFE F NSLEQLCIN+TNE LQ++FN
Sbjct 448 QQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCINFTNEKLQQFFN 493
> Hs10835119
Length=1855
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F+ NS EQ CINY NE LQ+ FN
Sbjct 432 FIGVLDIYGFETFEINSFEQFCINYANEKLQQQFN 466
> SPBC146.13c
Length=1217
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 21/32 (65%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
G+L+I+GFE F+ NS EQLCINY NE LQ+ F
Sbjct 411 GILDIYGFEIFENNSFEQLCINYVNEKLQQIF 442
> Hs16160857
Length=1855
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F+ NS EQ CINY NE LQ+ FN
Sbjct 432 FIGVLDIYGFETFEINSFEQFCINYANEKLQQQFN 466
> At2g20290
Length=1502
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 22/32 (68%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
GVL+I+GFE FK NS EQ CIN TNE LQ++F
Sbjct 440 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHF 471
> Hs11321579
Length=1938
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 33 EEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
++ + K+ F GVL+I GFE F NSLEQLCIN+TNE LQ++FN
Sbjct 447 QQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCINFTNEKLQQFFN 492
> Hs4505301
Length=1937
Score = 50.8 bits (120), Expect = 7e-07, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 33 EEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
++ + K+ F GVL+I GFE F NSLEQLCIN+TNE LQ++FN
Sbjct 448 QQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCINFTNEKLQQFFN 493
> Hs11024712
Length=1939
Score = 50.8 bits (120), Expect = 7e-07, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 33 EEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
++ + K+ F GVL+I GFE F NSLEQLCIN+TNE LQ++FN
Sbjct 448 QQLDTKQPRQYFIGVLDIAGFEIFDFNSLEQLCINFTNEKLQQFFN 493
> Hs11342672
Length=1940
Score = 50.4 bits (119), Expect = 7e-07, Method: Composition-based stats.
Identities = 25/35 (71%), Positives = 30/35 (85%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F+ NSLEQLCIN+TNE LQ++FN
Sbjct 457 FIGVLDIAGFEIFEYNSLEQLCINFTNEKLQQFFN 491
Lambda K H
0.311 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171925608
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40