bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
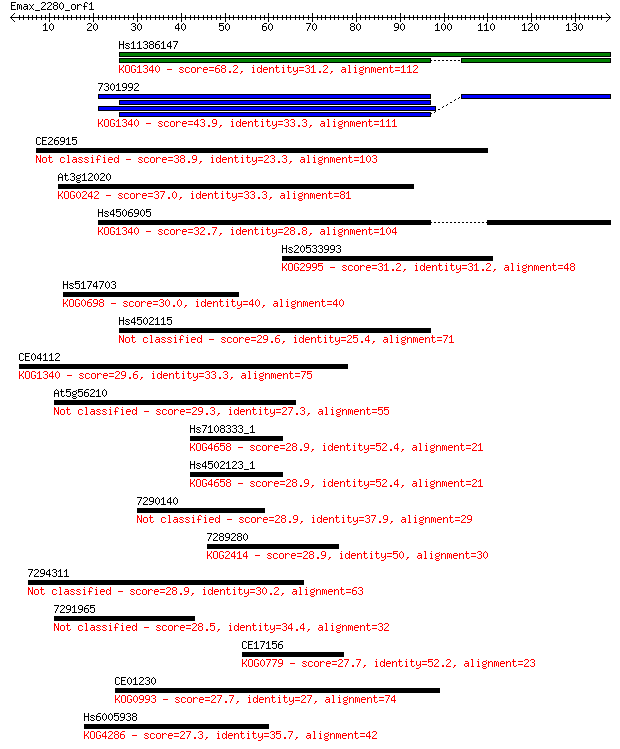
Query= Emax_2280_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
Hs11386147 68.2 4e-12
7301992 43.9 9e-05
CE26915 38.9 0.003
At3g12020 37.0 0.011
Hs4506905 32.7 0.22
Hs20533993 31.2 0.62
Hs5174703 30.0 1.3
Hs4502115 29.6 1.5
CE04112 29.6 1.9
At5g56210 29.3 2.2
Hs7108333_1 28.9 2.8
Hs4502123_1 28.9 2.8
7290140 28.9 2.9
7289280 28.9 3.2
7294311 28.9 3.3
7291965 28.5 4.0
CE17156 27.7 5.9
CE01230 27.7 7.4
Hs6005938 27.3 8.4
> Hs11386147
Length=524
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 56/115 (48%), Gaps = 4/115 (3%)
Query 26 CEMCQNIVNNLQ---QVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V L + S E+ E CS LP ++ +C + + P +++
Sbjct 409 CEVCKKLVGYLDRNLEKNSTKQEILAALEKGCSFLPDPYQKQCDQFVAEYEPVLIEILVE 468
Query 83 QDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+ +C I C + H LLG KC +GPSYWC + A CNA +C++ V
Sbjct 469 VMDPSFVCLKIGACPS-AHKPLLGTEKCIWGPSYWCQNTETAAQCNAVEHCKRHV 522
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+LG +CT G + WC V A C A +C Q V
Sbjct 19 VLGLKECTRGSAVWCQNVKTASDCGAVKHCLQTV 52
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/74 (20%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQV---QSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V + ++ + E+ + CSKLPK+ EC ++ + ++
Sbjct 315 CEVCEFLVKEVTKLIDNNKTEKEILDAFDKMCSKLPKSLSEECQEVVDTYGSSILSILLE 374
Query 83 QDNAKEICKSIDIC 96
+ + + +C + +C
Sbjct 375 EVSPELVCSMLHLC 388
> 7301992
Length=953
Score = 43.9 bits (102), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
LLG KCT+GPSYWC ++ C A +C Q V
Sbjct 25 LLGSSKCTWGPSYWCGNFSNSKECRATRHCIQTV 58
Score = 31.2 bits (69), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 4/80 (5%)
Query 21 NAEKRCEMCQNIVNNLQ-QVQSH--DLELDQLTENSCSKLP-KAHRTECGLMMKAFAPYF 76
+ + C +C+++V + Q++S+ + EL ++ E SC +P K + EC + F P
Sbjct 67 DTDSICTICKDMVTQARDQLKSNQTEEELKEVFEGSCKLIPIKPIQKECIKVADDFLPEL 126
Query 77 FDMMNHQDNAKEICKSIDIC 96
+ + Q N ++C +C
Sbjct 127 VEALASQMNPDQVCSVAGLC 146
Score = 31.2 bits (69), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQV---QSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
C +C+ +++ +Q+ S D E+ EN C+KLP +C ++ + ++
Sbjct 437 CTLCEYMLHFIQETLATPSTDDEIKHTVENICAKLPSGVAGQCRNFVEMYGDAVIALLVQ 496
Query 83 QDNAKEICKSIDIC 96
N +++C + +C
Sbjct 497 GLNPRDVCPLMQMC 510
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/80 (21%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Query 21 NAEKRCEMCQNIVNNLQQVQSHDLELDQLT---ENSCSKLPKAHRTECGLMMKAFAPYFF 77
A C +C+ +V L++ D + E SC ++ K T+C ++ +
Sbjct 661 TAAPNCLICEELVKTLEKRMGKHPTRDSIKHILEESCDRMRKPMNTKCHKVIDKYGDKIA 720
Query 78 DMMNHQDNAKEICKSIDICM 97
D++ + + K IC + +C+
Sbjct 721 DLLLKEMDPKLICTELGMCI 740
Score = 27.7 bits (60), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 36/74 (48%), Gaps = 4/74 (5%)
Query 26 CEMCQNIVNNLQQV---QSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V +L+ V + + E Q+ E C K K + EC ++ + ++ +
Sbjct 287 CELCEQLVKHLRDVLVANTTETEFKQVMEGFC-KQSKGFKDECLSIVDQYYHVIYETLVS 345
Query 83 QDNAKEICKSIDIC 96
+ +A C I IC
Sbjct 346 KLDANGACCMIGIC 359
> CE26915
Length=118
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 46/104 (44%), Gaps = 1/104 (0%)
Query 7 TKPNISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSK-LPKAHRTEC 65
T N+ K + + ++ C +C N+V L QV H +++ + C + +P C
Sbjct 15 TVSNVEGAKKMHSDTSKPLCGLCVNVVKQLDQVLEHGGDIEAAVDKFCKEDVPSFMVDMC 74
Query 66 GLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTPGHVHLLGGHK 109
+++ Y + + + A +IC I +C TP + L K
Sbjct 75 EKVIEKNLEYIINKLKDHEEADKICTDILLCRTPKQYYFLETQK 118
> At3g12020
Length=986
Score = 37.0 bits (84), Expect = 0.011, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 41/83 (49%), Gaps = 11/83 (13%)
Query 12 SDNKLI--VLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTECGLMM 69
+DN++I LN CE+ Q V NL+Q S LEL Q++ + R C L
Sbjct 739 ADNRIIQQTLNEKTCECEVLQEEVANLKQQLSEALELAQVSLFT-------FRKNCCLQK 791
Query 70 KAFAPYFFDMMNHQDNAKEICKS 92
F+P F + + +AKE+ +S
Sbjct 792 PCFSPIFIKEL--KQDAKELSES 812
> Hs4506905
Length=381
Score = 32.7 bits (73), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 110 CTFGPSYWCHTVDHAHACNAATYCRQKV 137
C GP +WC +++ A C A +C Q+V
Sbjct 32 CAQGPEFWCQSLEQALQCRALGHCLQEV 59
Score = 30.4 bits (67), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 21 NAEKRCEMCQNIVNNLQQVQSHDLELD---QLTENSCSKLP-KAHRTECGLMMKAFAPYF 76
A+ C+ C++IV+ L ++ + D + E C+ LP K +C ++ + P
Sbjct 64 GADDLCQECEDIVHILNKMAKEAIFQDTMRKFLEQECNVLPLKLLMPQCNQVLDDYFPLV 123
Query 77 FDMMNHQDNAKEICKSIDIC 96
D +Q ++ IC + +C
Sbjct 124 IDYFQNQIDSNGICMHLGLC 143
> Hs20533993
Length=441
Score = 31.2 bits (69), Expect = 0.62, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 63 TECGLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTPGHVHL--LGGHKC 110
T C +++A A +F + NH +C +++I + H HL GG++C
Sbjct 167 TLCKALLQAHAHFFPECSNHNTQDLRVCLNLEIGLRKYHPHLKKFGGNRC 216
> Hs5174703
Length=504
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 3/40 (7%)
Query 13 DNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTEN 52
+NKL V N R +C++ V+ LQ Q L +D TEN
Sbjct 175 NNKLYVANVGTNRALLCKSTVDGLQVTQ---LNVDHTTEN 211
> Hs4502115
Length=575
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 35/73 (47%), Gaps = 4/73 (5%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAH--RTECGLMMKAFAPYFFDMMNHQ 83
C + +++ L QV H+ + E CS LP+ +T C L++ F +++
Sbjct 44 CVLVVSVIEQLAQV--HNSTVQASMERLCSYLPEKLFLKTTCYLVIDKFGSDIIKLLSAD 101
Query 84 DNAKEICKSIDIC 96
NA +C +++ C
Sbjct 102 MNADVVCHTLEFC 114
> CE04112
Length=402
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 36/80 (45%), Gaps = 8/80 (10%)
Query 3 IHVCTKPNISDNKLIVLNNAEK---RCEMCQNIVNNLQQVQS--HDLELDQLTENSCSKL 57
+H C K +N L+ + +EK CE C+ + + Q Q H +D L N C KL
Sbjct 309 MHSCEKK---ENALVEMAMSEKVMLGCENCKAVEHFFAQNQEALHSHAVDGLYSNVCQKL 365
Query 58 PKAHRTECGLMMKAFAPYFF 77
P A T C + + FF
Sbjct 366 PTALGTMCEASIIRLSRKFF 385
> At5g56210
Length=509
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 11 ISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTEC 65
I + K + L+ +K +N+ N Q +Q+H +E+ ++ E C K T C
Sbjct 430 IEEEKTLGLSKLDKAETKAENLKNQAQDLQNHCVEITEIQEVECLKKRAFKTTRC 484
> Hs7108333_1
Length=569
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 42 HDLELDQLTENSCSKLPKAHR 62
HDL++D LTE +CS+L H+
Sbjct 427 HDLQVDFLTEKNCSQLQDLHK 447
> Hs4502123_1
Length=569
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 42 HDLELDQLTENSCSKLPKAHR 62
HDL++D LTE +CS+L H+
Sbjct 427 HDLQVDFLTEKNCSQLQDLHK 447
> 7290140
Length=1279
Score = 28.9 bits (63), Expect = 2.9, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 30 QNIVNNLQQVQSHDLELDQLTENSCSKLP 58
Q +++NLQ++Q ++ LDQ+T + S P
Sbjct 1096 QRLISNLQELQQLNVSLDQVTTATSSPRP 1124
> 7289280
Length=545
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Query 46 LDQLTENSCSKLPKAHRTECGLMMKAFAPY 75
LDQL E +C KL K + E GL+ K+F+ Y
Sbjct 405 LDQLFETTCYKLGK-YLQEIGLVGKSFSEY 433
> 7294311
Length=1188
Score = 28.9 bits (63), Expect = 3.3, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Query 5 VCTKPNISDNKLIVLNNAEKRCEMCQNI-VNNLQQVQSHDLELDQLTENSCSKLPKAHRT 63
V K D+KL +L A KRC + + +VQ + +LD + +++ KLP
Sbjct 731 VSQKVQKKDDKLNMLKQATKRCRKLRALNAKRKNEVQKLEQDLDTIRKSAPHKLPIIRLK 790
Query 64 ECGL 67
C L
Sbjct 791 RCNL 794
> 7291965
Length=345
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 11 ISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSH 42
IS+N +I ++ E+ C +C+ +V + Q+Q+H
Sbjct 231 ISENSVIKSDDNERTCHLCKIVVTSAAQMQAH 262
> CE17156
Length=311
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 1/24 (4%)
Query 54 CSKLPK-AHRTECGLMMKAFAPYF 76
C KLP+ + +CG+ M AFA YF
Sbjct 242 CQKLPQQKNSVDCGIFMMAFAEYF 265
> CE01230
Length=414
Score = 27.7 bits (60), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 15/74 (20%)
Query 25 RCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNHQD 84
+CEMCQN NL +Q ++ +L + E M+ Y D+ N +D
Sbjct 36 QCEMCQNYEVNLTALQENERKL---------------KEEVKAAMELNERYQLDLSNERD 80
Query 85 NAKEICKSIDICMT 98
KE+ K ++ T
Sbjct 81 YRKELEKKMNELST 94
> Hs6005938
Length=3433
Score = 27.3 bits (59), Expect = 8.4, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 27/45 (60%), Gaps = 3/45 (6%)
Query 18 VLNNAEKRCEMCQNIVNNLQQVQSHDLE---LDQLTENSCSKLPK 59
VLN++E + ++ N++N+L +V+ E LD++ EN L K
Sbjct 922 VLNDSENKAQVSLNVLNDLAKVEKALQEKKTLDEILENQKPALHK 966
Lambda K H
0.323 0.133 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40