bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2277_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
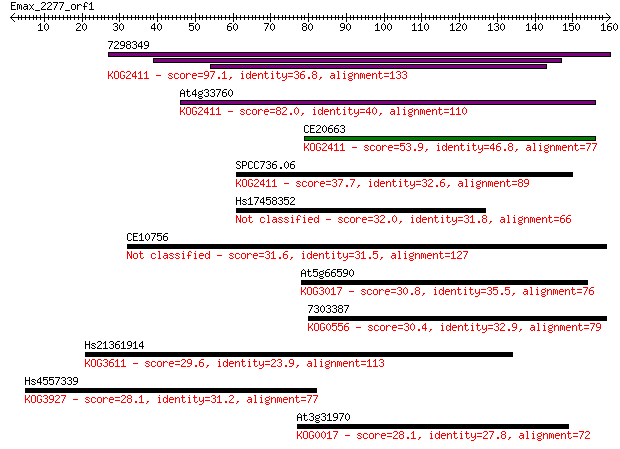
7298349 97.1 1e-20
At4g33760 82.0 5e-16
CE20663 53.9 1e-07
SPCC736.06 37.7 0.010
Hs17458352 32.0 0.57
CE10756 31.6 0.70
At5g66590 30.8 1.2
7303387 30.4 1.4
Hs21361914 29.6 2.3
Hs4557339 28.1 7.4
At3g31970 28.1 7.5
> 7298349
Length=1052
Score = 97.1 bits (240), Expect = 1e-20, Method: Composition-based stats.
Identities = 49/136 (36%), Positives = 78/136 (57%), Gaps = 3/136 (2%)
Query 27 STLNPQHTANSVKAECDVNRFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDS 86
+T N A D N+F R+H CG++ + + E+V +CGW+E+ R KF LRD+
Sbjct 352 TTANASLIAEQRAKVADTNKFADRTHNCGELTSNDINEKVVICGWLEFQRMNKFFILRDA 411
Query 87 YGSVQLFV--KSSELQEIVRN-LSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELS 143
YG Q+ + K+ L+E + +ES+++ +G V RP +N K+ G+VEV +++
Sbjct 412 YGQTQVLLSPKTYGLEEYAETGVPIESIVRVEGTVIPRPAATINPKMQTGHVEVEADKVV 471
Query 144 VLNMCKANLPFLNRDY 159
VLN K NLPF R +
Sbjct 472 VLNPAKKNLPFEIRKF 487
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 33/108 (30%), Positives = 41/108 (37%), Gaps = 30/108 (27%)
Query 39 KAECDVNRFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYGSVQLFVKSSE 98
K E + F R C + VG+ VTL GWI + KFL L+D YG QL ++
Sbjct 223 KNENILKYFENRDLTCNDLRRDDVGKTVTLVGWIPSTKNNKFLQLKDGYGQTQLMIED-- 280
Query 99 LQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLN 146
SK G VEVSV + VLN
Sbjct 281 ----------------------------QSKYDTGEVEVSVTSVKVLN 300
Score = 35.8 bits (81), Expect = 0.040, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 43/93 (46%), Gaps = 4/93 (4%)
Query 54 CGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRD-SYGSVQLFVKSSELQEIVR---NLSLE 109
CG++ + G V L G + R +F LRD + G+ QL V + + R N+
Sbjct 91 CGELRNHHCGLFVELSGRLIKKRVNRFAELRDRNGGACQLVVLEDKHPRVARRMNNMPEN 150
Query 110 SVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEEL 142
+ + G V RP N + G +EV V+++
Sbjct 151 TTLTIVGLVMRRPHNSCNQTMPTGEIEVEVQDI 183
> At4g33760
Length=636
Score = 82.0 bits (201), Expect = 5e-16, Method: Composition-based stats.
Identities = 44/116 (37%), Positives = 64/116 (55%), Gaps = 6/116 (5%)
Query 46 RFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRF---CKFLTLRDSYGSVQLFVKSSELQE- 101
R+ R+ CG++ + VG+ V LCGW+ HR FL LRD G VQ+ E E
Sbjct 71 RWVSRTELCGELSVNDVGKRVHLCGWVALHRVHGGLTFLNLRDHTGIVQVRTLPDEFPEA 130
Query 102 --IVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKANLPFL 155
++ ++ LE V+ +G V RP++ VN K+ G VEV E + +LN + LPFL
Sbjct 131 HGLINDMRLEYVVLVEGTVRSRPNESVNKKMKTGFVEVVAEHVEILNPVRTKLPFL 186
> CE20663
Length=569
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 55/104 (52%), Gaps = 28/104 (26%)
Query 79 KFLTLRDSYGSVQLFVK-SSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEV 137
+F LRD+YGSVQ + SS+LQ +++++ ESV++ G V R D + NSK+ G++EV
Sbjct 3 RFFVLRDAYGSVQAKISASSKLQSLLKDIPYESVVRVDGIVVDRGD-NRNSKMKTGDIEV 61
Query 138 S--------------------------VEELSVLNMCKANLPFL 155
+ VEEL+VLN +N+P L
Sbjct 62 NFQKYYFKSGNNENSNREVFDFLILIDVEELTVLNKATSNIPML 105
> SPCC736.06
Length=611
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 54/117 (46%), Gaps = 31/117 (26%)
Query 61 LVGEEVTLCGWI-----EYHRFCKFLTLRDSYGSV-QLFV-------------------- 94
LVG +V++ GW+ + + F LRD++G++ QL
Sbjct 13 LVGTKVSIQGWLVATSRQVSKSISFHQLRDTHGTILQLLSTDKIILQQKREPLVSSTDFS 72
Query 95 --KSSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCK 149
KS+ + + ++ ESV++ GK+ RP+ D + E+ VE++ +LN+ K
Sbjct 73 QQKSTSVMRTLSSIPPESVVQVTGKLQRRPEHD---RRPGNEFELHVEDVKLLNVAK 126
> Hs17458352
Length=388
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 31/67 (46%), Gaps = 4/67 (5%)
Query 61 LVGEEVTLCGWIEY-HRFCKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKVN 119
L+ + + WIEY H F L SY Q + K LQ R +SLE ++ +
Sbjct 60 LLNRTLAVPPWIEYQHHKPPFTNLHVSY---QKYFKLEPLQAYHRVISLEDFMEKLAPTH 116
Query 120 YRPDKDV 126
+ P+K V
Sbjct 117 WPPEKRV 123
> CE10756
Length=276
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 62/132 (46%), Gaps = 18/132 (13%)
Query 32 QHTANSVKAECDVNRFTYRSHCCGQVDDSLVGEEVTLCG-WIEYHRF---CKFLTLRDSY 87
Q ++N + ECDV+ RS C Q+ EE+ L +E R K+L +
Sbjct 5 QTSSNDICQECDVSEIGQRSSNC-QIASIRANEEILLRAIQLEDQRINDSKKYLFSTHTR 63
Query 88 GSVQLFVKSSE-LQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLN 146
+Q F K+ E L +++RNL+ E IK + + P +V +V+G VE
Sbjct 64 EVIQKFHKTFEPLDDVLRNLN-EIYIKCIPEAGFFP--EVKKGVVDGFVE---------K 111
Query 147 MCKANLPFLNRD 158
+ ANL F NR+
Sbjct 112 IADANLSFKNRN 123
> At5g66590
Length=185
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 17/91 (18%)
Query 78 CKFLTLRDS-YGSVQLFVKSSELQEIVRNLSLESVIKDKGKVNYRPD-----------KD 125
C+F +L YG+ QL+ K L + +L++E+ +K+K NY+ D K
Sbjct 88 CEFASLNPGKYGANQLWAKG--LVAVTPSLAVETWVKEKPFYNYKSDTCAANHTCGVYKQ 145
Query 126 V---NSKLVNGNVEVSVEELSVLNMCKANLP 153
V NSK + +E +VL +C N P
Sbjct 146 VVWRNSKELGCAQATCTKESTVLTICFYNPP 176
> 7303387
Length=531
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 42/87 (48%), Gaps = 13/87 (14%)
Query 80 FLTLRDSYGSVQLF-----VKSSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKL---V 131
FL LR +VQ V S ++ + N+ ES+I + K V+SK+
Sbjct 106 FLILRQQSSTVQCILAVGDVISKQMVKFAGNIPKESIIDIQAK-----PVAVSSKIESCT 160
Query 132 NGNVEVSVEELSVLNMCKANLPFLNRD 158
++E+SVE++ V++ KA LP D
Sbjct 161 EQSLELSVEQIFVISQAKAQLPLQIED 187
> Hs21361914
Length=761
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 27/116 (23%), Positives = 47/116 (40%), Gaps = 19/116 (16%)
Query 21 PQCCFLSTLNPQHTANSVKAECDVNRFTYRSHCCGQVDDSLVGEEVT---LCGWIEYHRF 77
P L + P ++ K+EC + + + C + LV VT CG +
Sbjct 92 PGVPRLKNMIPWPASDRKKSECAFKKKSNETQCFNFIR-VLVSYNVTHLYTCGTFAFSPA 150
Query 78 CKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNG 133
C F+ L+DSY + +S + V++ KG+ + P + LV+G
Sbjct 151 CTFIELQDSY---------------LLPISEDKVMEGKGQSPFDPAHKHTAVLVDG 191
> Hs4557339
Length=291
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 34/78 (43%), Gaps = 6/78 (7%)
Query 5 SSVNPASQALSRNHSLPQCCFLSTLN-PQHTANSVKAECDVNRFTYRSHCCGQVDDSLVG 63
+ +PA+Q S N + Q F + P HT S K D+ +C G D +
Sbjct 118 AGYSPAAQEDSINCTSEQYFFQESFRAPNHTKFSCKFTADM-----LQNCSGLADPNFGF 172
Query 64 EEVTLCGWIEYHRFCKFL 81
EE C I+ +R KFL
Sbjct 173 EEGKPCFIIKMNRIVKFL 190
> At3g31970
Length=1329
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 34/72 (47%), Gaps = 2/72 (2%)
Query 77 FCKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVE 136
F +L+ + +L ++ E+V + LE + GK N D ++ K V G+VE
Sbjct 769 FTDHKSLKYIFTQPELNLRQRRWMELVADYDLE-IAYHSGKANVVADA-LSRKRVGGSVE 826
Query 137 VSVEELSVLNMC 148
V E+ L +C
Sbjct 827 ALVSEIGALRLC 838
Lambda K H
0.318 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40