bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2230_orf2
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
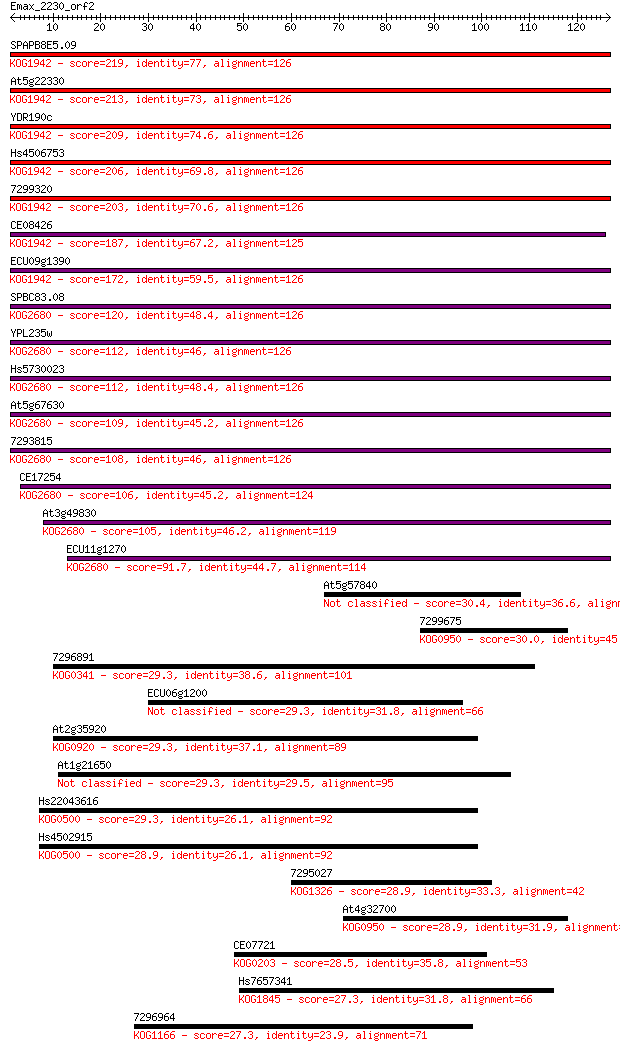
SPAPB8E5.09 219 9e-58
At5g22330 213 9e-56
YDR190c 209 1e-54
Hs4506753 206 6e-54
7299320 203 6e-53
CE08426 187 4e-48
ECU09g1390 172 1e-43
SPBC83.08 120 8e-28
YPL235w 112 1e-25
Hs5730023 112 2e-25
At5g67630 109 1e-24
7293815 108 2e-24
CE17254 106 1e-23
At3g49830 105 3e-23
ECU11g1270 91.7 3e-19
At5g57840 30.4 0.93
7299675 30.0 1.2
7296891 29.3 1.7
ECU06g1200 29.3 1.7
At2g35920 29.3 1.7
At1g21650 29.3 1.8
Hs22043616 29.3 2.1
Hs4502915 28.9 2.1
7295027 28.9 2.2
At4g32700 28.9 2.3
CE07721 28.5 2.9
Hs7657341 27.3 6.8
7296964 27.3 6.9
> SPAPB8E5.09
Length=456
Score = 219 bits (558), Expect = 9e-58, Method: Compositional matrix adjust.
Identities = 97/126 (76%), Positives = 118/126 (93%), Gaps = 0/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEANTG VKRVGRSDA+ATEFDLEAE++VP+PKG+V KRKE+VQ V+LHDLD+AN++P
Sbjct 192 IYIEANTGAVKRVGRSDAYATEFDLEAEEYVPMPKGEVHKRKEIVQDVTLHDLDIANARP 251
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QGG D++SMMG MKP+KTEIT++LR EINK VNKYIEQG+A+LIPGVLFIDEVHMLDIE
Sbjct 252 QGGQDIMSMMGQLMKPKKTEITDKLRGEINKVVNKYIEQGIAELIPGVLFIDEVHMLDIE 311
Query 121 CFSFLN 126
CF++LN
Sbjct 312 CFTYLN 317
> At5g22330
Length=458
Score = 213 bits (541), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 92/126 (73%), Positives = 117/126 (92%), Gaps = 0/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEAN+G VKRVGRSDAFATEFDLEAE++VP+PKG+V K+KE+VQ V+L DLD AN++P
Sbjct 194 IYIEANSGAVKRVGRSDAFATEFDLEAEEYVPLPKGEVHKKKEIVQDVTLQDLDAANARP 253
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QGG D++S+MG MKPRKTEIT++LR+EINK VN+YI++GVA+L+PGVLFIDEVHMLD+E
Sbjct 254 QGGQDILSLMGQMMKPRKTEITDKLRQEINKVVNRYIDEGVAELVPGVLFIDEVHMLDME 313
Query 121 CFSFLN 126
CFS+LN
Sbjct 314 CFSYLN 319
> YDR190c
Length=463
Score = 209 bits (532), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 94/126 (74%), Positives = 116/126 (92%), Gaps = 0/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEANTG VKRVGRSDA+ATEFDLE E++VP+PKG+V K+KE+VQ V+LHDLDVAN++P
Sbjct 200 IYIEANTGAVKRVGRSDAYATEFDLETEEYVPLPKGEVHKKKEIVQDVTLHDLDVANARP 259
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QGG DVISMMG +KP+KTEITE+LR+E+NK V KYI+QGVA+LIPGVLFIDEV+MLDIE
Sbjct 260 QGGQDVISMMGQLLKPKKTEITEKLRQEVNKVVAKYIDQGVAELIPGVLFIDEVNMLDIE 319
Query 121 CFSFLN 126
F++LN
Sbjct 320 IFTYLN 325
> Hs4506753
Length=456
Score = 206 bits (525), Expect = 6e-54, Method: Compositional matrix adjust.
Identities = 88/126 (69%), Positives = 114/126 (90%), Gaps = 0/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEAN+G VKR GR D +ATEFDLEAE++VP+PKG V K+KE++Q V+LHDLDVAN++P
Sbjct 191 IYIEANSGAVKRQGRCDTYATEFDLEAEEYVPLPKGDVHKKKEIIQDVTLHDLDVANARP 250
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QGG D++SMMG MKP+KTEIT++LR EINK VNKYI+QG+A+L+PGVLF+DEVHMLDIE
Sbjct 251 QGGQDILSMMGQLMKPKKTEITDKLRGEINKVVNKYIDQGIAELVPGVLFVDEVHMLDIE 310
Query 121 CFSFLN 126
CF++L+
Sbjct 311 CFTYLH 316
> 7299320
Length=456
Score = 203 bits (517), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 89/126 (70%), Positives = 113/126 (89%), Gaps = 0/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEAN+G VKR GRSD FATEFDLE E++VP+PKG V K+KEV+Q V+LHDLDVAN++P
Sbjct 191 IYIEANSGAVKRQGRSDTFATEFDLETEEYVPLPKGDVHKKKEVIQDVTLHDLDVANARP 250
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QGG DV+SMMG MKP+KTEIT++LR EINK VNKYI+QG+A+L+PGVLFIDE+HMLD+E
Sbjct 251 QGGQDVLSMMGQLMKPKKTEITDKLRMEINKVVNKYIDQGIAELVPGVLFIDEIHMLDLE 310
Query 121 CFSFLN 126
F++L+
Sbjct 311 TFTYLH 316
> CE08426
Length=458
Score = 187 bits (475), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 84/126 (66%), Positives = 109/126 (86%), Gaps = 1/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++IEAN+GIVKRVGR D +A+EFDLEA++FVP+PKG V K K++VQ VSLHDLD+AN++P
Sbjct 209 IYIEANSGIVKRVGRCDVYASEFDLEADEFVPMPKGDVRKSKDIVQNVSLHDLDIANARP 268
Query 61 QGGT-DVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDI 119
QG DV +++ M P+KTE+T+RLR EINK VN+YIE GVA+L+PGVLFIDEVHMLD+
Sbjct 269 QGRQGDVSNIVSQLMTPKKTEVTDRLRSEINKVVNEYIESGVAELMPGVLFIDEVHMLDV 328
Query 120 ECFSFL 125
ECF++L
Sbjct 329 ECFTYL 334
> ECU09g1390
Length=426
Score = 172 bits (437), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 75/126 (59%), Positives = 108/126 (85%), Gaps = 1/126 (0%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
V+IE N+G++K++GRS+A +FDLEA+ +VP+PKG+V KRKEV+Q+V+LHDLD+AN++P
Sbjct 183 VYIEVNSGVIKKLGRSEAHMNDFDLEADTYVPIPKGEVLKRKEVMQSVTLHDLDMANARP 242
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
G D++S++ + PRKTEITERLR ++N+ VN Y+E G A+++PGVLFIDEVHMLD+E
Sbjct 243 SG-QDMLSLVFRILSPRKTEITERLRGDVNRMVNGYLENGNAEIVPGVLFIDEVHMLDVE 301
Query 121 CFSFLN 126
CF+FL+
Sbjct 302 CFTFLH 307
> SPBC83.08
Length=465
Score = 120 bits (300), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 61/128 (47%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANS 58
+ I+ + G V ++GRS + A ++D D FV P+G+++KRKEVV TVSLHD+DV NS
Sbjct 187 ISIDKSVGRVTKLGRSFSRARDYDAMGADTRFVQCPQGEIQKRKEVVHTVSLHDIDVINS 246
Query 59 KPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLD 118
+ QG + S +KP +RE+IN V+++ E+G A+++PGVLF+DEVHMLD
Sbjct 247 RTQGFLALFSGDTGEIKPE-------VREQINTKVSEWREEGKAEIVPGVLFVDEVHMLD 299
Query 119 IECFSFLN 126
IECFSF N
Sbjct 300 IECFSFFN 307
> YPL235w
Length=471
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 84/128 (65%), Gaps = 9/128 (7%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANS 58
+ I+ +G + ++GRS A + ++D D FV P+G+++KRK VV TVSLH++DV NS
Sbjct 190 ISIDKASGKITKLGRSFARSRDYDAMGADTRFVQCPEGELQKRKTVVHTVSLHEIDVINS 249
Query 59 KPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLD 118
+ QG A EI +R++IN V ++ E+G A+++PGVLFIDEVHMLD
Sbjct 250 RTQG-------FLALFTGDTGEIRSEVRDQINTKVAEWKEEGKAEIVPGVLFIDEVHMLD 302
Query 119 IECFSFLN 126
IECFSF+N
Sbjct 303 IECFSFIN 310
> Hs5730023
Length=463
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 61/128 (47%), Positives = 81/128 (63%), Gaps = 9/128 (7%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANS 58
+ I+ TG + ++GRS A ++D FV P G+++KRKEVV TVSLH++DV NS
Sbjct 193 ITIDKATGKISKLGRSFTRARDYDAMGSQTKFVQCPDGELQKRKEVVHTVSLHEIDVINS 252
Query 59 KPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLD 118
+ QG A EI +RE+IN V ++ E+G A++IPGVLFIDEVHMLD
Sbjct 253 RTQG-------FLALFSGDTGEIKSEVREQINAKVAEWREEGKAEIIPGVLFIDEVHMLD 305
Query 119 IECFSFLN 126
IE FSFLN
Sbjct 306 IESFSFLN 313
> At5g67630
Length=469
Score = 109 bits (273), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/128 (44%), Positives = 83/128 (64%), Gaps = 9/128 (7%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANS 58
+ I+ TG + ++GRS + + ++D FV P+G+++KRKEVV V+LH++DV NS
Sbjct 190 IAIDKATGKITKLGRSFSRSRDYDAMGAQTKFVQCPEGELQKRKEVVHCVTLHEIDVINS 249
Query 59 KPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLD 118
+ QG A EI +RE+I+ V ++ E+G A+++PGVLFIDEVHMLD
Sbjct 250 RTQG-------FLALFTGDTGEIRSEVREQIDTKVAEWREEGKAEIVPGVLFIDEVHMLD 302
Query 119 IECFSFLN 126
IECFSFLN
Sbjct 303 IECFSFLN 310
> 7293815
Length=481
Score = 108 bits (270), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 82/128 (64%), Gaps = 9/128 (7%)
Query 1 VFIEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANS 58
+ I+ +G V ++GRS A ++D FV P+G+++KRKEVV TV+LH++DV NS
Sbjct 189 ITIDKASGKVNKLGRSFTRARDYDATGAQTRFVQCPEGELQKRKEVVHTVTLHEIDVINS 248
Query 59 KPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLD 118
+ G A EI + +R++IN V ++ E+G A++ PGVLFIDEVHMLD
Sbjct 249 RTHG-------FLALFSGDTGEIKQEVRDQINNKVLEWREEGKAEINPGVLFIDEVHMLD 301
Query 119 IECFSFLN 126
IECFSFLN
Sbjct 302 IECFSFLN 309
> CE17254
Length=448
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/126 (44%), Positives = 77/126 (61%), Gaps = 9/126 (7%)
Query 3 IEANTGIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANSKP 60
++ +G V R+GRS + ++D V P G+++KR+E V TV LHD+DV NS+
Sbjct 191 VDKASGRVTRLGRSFNRSHDYDAMGPKVKLVQCPDGEIQKRRETVHTVCLHDIDVINSRT 250
Query 61 QGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 120
QG A EI +R++INK V ++ E+G A+ +PGVLFIDE HMLDIE
Sbjct 251 QGYV-------ALFSGDTGEIKAEVRDQINKKVLEWREEGKAKFVPGVLFIDEAHMLDIE 303
Query 121 CFSFLN 126
CFSFLN
Sbjct 304 CFSFLN 309
> At3g49830
Length=473
Score = 105 bits (261), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 55/121 (45%), Positives = 78/121 (64%), Gaps = 9/121 (7%)
Query 8 GIVKRVGRSDAFATEFDLEAED--FVPVPKGQVEKRKEVVQTVSLHDLDVANSKPQGGTD 65
G + ++GRS + +FD+ FV P+G++EKRKEV+ +V+LH++DV NS+ QG
Sbjct 198 GKITKLGRSFTRSRDFDVMGSKTKFVQCPEGELEKRKEVLHSVTLHEIDVINSRTQGYL- 256
Query 66 VISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFL 125
A EI RE+ + V ++ E+G A+++PGVLFIDEVHMLDIECFSFL
Sbjct 257 ------ALFTGDTGEIRSETREQSDTKVAEWREEGKAEIVPGVLFIDEVHMLDIECFSFL 310
Query 126 N 126
N
Sbjct 311 N 311
> ECU11g1270
Length=418
Score = 91.7 bits (226), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 51/114 (44%), Positives = 67/114 (58%), Gaps = 12/114 (10%)
Query 13 VGRSDAFATEFDLEAEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKPQGGTDVISMMGA 72
V RSD T+ FVP P+G++ + E Q +SLHD+DV NSK +G A
Sbjct 186 VKRSDVVGTD-----TRFVPCPEGELIRITEETQEISLHDIDVVNSKAEGYL-------A 233
Query 73 YMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLN 126
EI R+E+NK V +I +G A+++ GVLFIDEVHMLDIE F+FLN
Sbjct 234 LFSGETGEIRAETRDEVNKKVWGWINEGKAEIVRGVLFIDEVHMLDIESFAFLN 287
> At5g57840
Length=443
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 9/50 (18%)
Query 67 ISMMGAYMKPRKTEITERLREEINKTVNKYIEQGV---------AQLIPG 107
+S +GA+ + +E ER+ EI K N+Y+ + QL+PG
Sbjct 310 VSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLERHPDLDQLVPG 359
> 7299675
Length=2059
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Query 87 EEINKTVNKYIEQGVAQLIPGVLFIDEVHML 117
E+ N VNK +EQG + I G++ +DEVH++
Sbjct 333 EKANSIVNKLMEQGKLETI-GMVVVDEVHLI 362
> 7296891
Length=619
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 61/132 (46%), Gaps = 32/132 (24%)
Query 10 VKRVGRSDAFATEFDLEAEDFVP-VP-----KGQVEKRKEVVQTVSLHDLDVANSK---- 59
VKR RS + E DL+ ED+VP VP K + K +VQ VS ++S+
Sbjct 4 VKRYRRSSKSSEEGDLDNEDYVPYVPVKERKKQHMIKLGRIVQLVSETAQPKSSSENENE 63
Query 60 --PQGGTDV--------ISMMGAYMKPRK-------TEITERLREEINKTVNKYIEQ--- 99
QG DV IS++ + + +K + + ++LREE K + +Q
Sbjct 64 DDSQGAHDVETWGRKYNISLLDQHTELKKIAEAKKLSAVEKQLREE-EKIMESIAQQKAL 122
Query 100 -GVAQLIPGVLF 110
GVA+L G+ +
Sbjct 123 MGVAELAKGIQY 134
> ECU06g1200
Length=541
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 2/68 (2%)
Query 30 FVPVPKGQVEKRKEVVQTV--SLHDLDVANSKPQGGTDVISMMGAYMKPRKTEITERLRE 87
F+ KG ++ KEVVQ S+++LD N Q ++ G +K E+ ERL++
Sbjct 450 FLNSNKGVTDEFKEVVQNFRNSVNELDFNNVTVQQLKSLMKEFGLNHTGKKNELIERLQD 509
Query 88 EINKTVNK 95
+ K K
Sbjct 510 ILKKIDGK 517
> At2g35920
Length=993
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 48/116 (41%), Gaps = 31/116 (26%)
Query 10 VKRVGRSDAFATEFDLEAEDFVPVPKGQVEKR-----------KEVVQTVSLHDLDVANS 58
KR G+ AF T+ +L D P G V R E V+T S++ D N
Sbjct 856 CKRRGKRTAFYTK-ELGKVDIHP---GSVNARVNLFSLPYLVYSEKVKTTSVYIRDSTNI 911
Query 59 K------------PQGGTDVISMMGAYMKPRKT----EITERLREEINKTVNKYIE 98
P + I M+G Y+ + E+ +RLR E++K +NK IE
Sbjct 912 SDYALLMFGGNLIPSKTGEGIEMLGGYLHFSASKNILELIQRLRGEVDKLLNKKIE 967
> At1g21650
Length=1579
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 45/101 (44%), Gaps = 15/101 (14%)
Query 11 KRVGRSDAFATEFDLEAEDFVPV---PKGQVEKRKEVVQTVS---LHDLDVANSKPQGGT 64
+R+ R ++ A D++AE F V K + R VQ + LHD +A K G
Sbjct 765 ERLARGESLA---DMQAEAFAVVREAAKRTIGMRHFDVQIIGGGVLHDGSIAEMKTGEGK 821
Query 65 DVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLI 105
++S + AY+ E + TVN Y+ Q A+ +
Sbjct 822 TLVSTLAAYLNALTGEGVHVV------TVNDYLAQRDAEWM 856
> Hs22043616
Length=686
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 47/107 (43%), Gaps = 16/107 (14%)
Query 7 TGIVKRVGRSDAFATEFDLEAEDFVPVPKGQV---EKRKEVVQTVSLHDLDVAN------ 57
T +K +G SD F D E P + EK K+++ L DL++AN
Sbjct 558 TANIKSIGYSDLFCLSKDDLMEALTEYPDAKTMLEEKGKQILMKDGLLDLNIANAGSDPK 617
Query 58 ------SKPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIE 98
++ +G D++ A + + ++L++ + K V K+++
Sbjct 618 DLEEKVTRMEGSVDLLQTRFARILAEYESMQQKLKQRLTK-VEKFLK 663
> Hs4502915
Length=690
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 47/107 (43%), Gaps = 16/107 (14%)
Query 7 TGIVKRVGRSDAFATEFDLEAEDFVPVPKGQV---EKRKEVVQTVSLHDLDVAN------ 57
T +K +G SD F D E P + EK K+++ L DL++AN
Sbjct 562 TANIKSIGYSDLFCLSKDDLMEALTEYPDAKTMLEEKGKQILMKDGLLDLNIANAGSDPK 621
Query 58 ------SKPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIE 98
++ +G D++ A + + ++L++ + K V K+++
Sbjct 622 DLEEKVTRMEGSVDLLQTRFARILAEYESMQQKLKQRLTK-VEKFLK 667
> 7295027
Length=1782
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 60 PQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGV 101
P GG D++ M + PR + L E + V Y+ QGV
Sbjct 1099 PGGGGDMVRNMSNPIHPRHQSTLQSLGETVKLLVRVYVVQGV 1140
> At4g32700
Length=1548
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Query 71 GAYMKPRKTEITERLREEINKTVNKYIEQG-VAQLIPGVLFIDEVHML 117
G P+ T + E+ N +N+ +E+G +++L G++ IDE+HM+
Sbjct 48 GGGTLPKDTSVAVCTIEKANSLINRLLEEGRLSEL--GIIVIDELHMV 93
> CE07721
Length=996
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 10/58 (17%)
Query 48 VSLHD-----LDVANSKPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQG 100
VS+HD L V P+ DV S + ++ +++E+T++LRE+ N Y+E G
Sbjct 467 VSIHDNGDHYLLVMKGAPERILDVCSTI--FLNGKESELTDKLREDFN---TAYLELG 519
> Hs7657341
Length=984
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 49 SLHDLDVANSKPQGGTDVISMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGV 108
S H ++ S P G IS P K E ++LRE + K N+ EQ Q P
Sbjct 522 SCHQVECLPSIPLGTMSTIS-------PSKNEKEKQLRESVIKYQNRLAEQ---QPQPQF 571
Query 109 LFIDEV 114
+ +DE+
Sbjct 572 IPVDEI 577
> 7296964
Length=1099
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 17/80 (21%), Positives = 37/80 (46%), Gaps = 9/80 (11%)
Query 27 AEDFVPVPKGQVEKRKEVVQTVSLHDLDVANSKPQ---------GGTDVISMMGAYMKPR 77
A D P G+ K ++VVQT +++ + T + + G YM+P+
Sbjct 969 AIDMTLFPDGEKTKFRKVVQTDGFTCIEMQEGRSWSYETDLFCIAATVHVMLFGDYMQPQ 1028
Query 78 KTEITERLREEINKTVNKYI 97
K + +R+++ + + K++
Sbjct 1029 KKGSSWEIRQKLPRYLKKHV 1048
Lambda K H
0.318 0.137 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40