bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
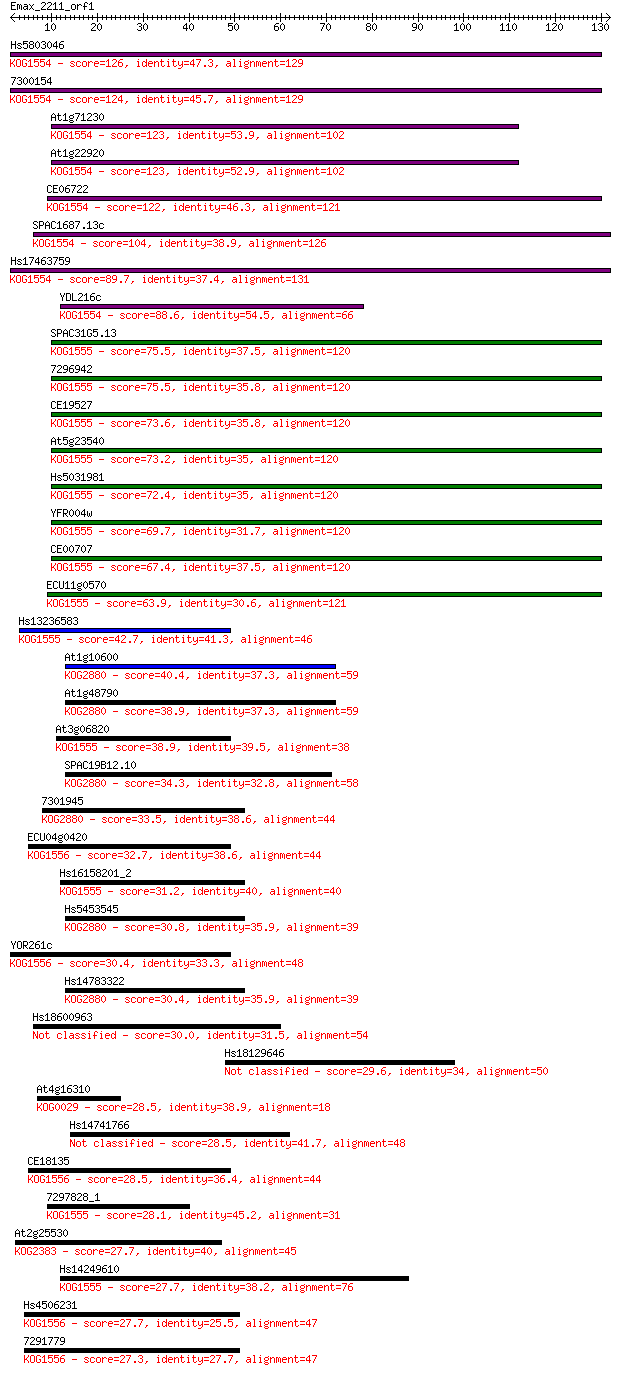
Query= Emax_2211_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
Hs5803046 126 1e-29
7300154 124 5e-29
At1g71230 123 1e-28
At1g22920 123 1e-28
CE06722 122 2e-28
SPAC1687.13c 104 3e-23
Hs17463759 89.7 1e-18
YDL216c 88.6 3e-18
SPAC31G5.13 75.5 2e-14
7296942 75.5 3e-14
CE19527 73.6 1e-13
At5g23540 73.2 1e-13
Hs5031981 72.4 2e-13
YFR004w 69.7 1e-12
CE00707 67.4 7e-12
ECU11g0570 63.9 8e-11
Hs13236583 42.7 2e-04
At1g10600 40.4 0.001
At1g48790 38.9 0.002
At3g06820 38.9 0.003
SPAC19B12.10 34.3 0.065
7301945 33.5 0.097
ECU04g0420 32.7 0.20
Hs16158201_2 31.2 0.60
Hs5453545 30.8 0.71
YOR261c 30.4 0.90
Hs14783322 30.4 0.94
Hs18600963 30.0 1.1
Hs18129646 29.6 1.7
At4g16310 28.5 3.3
Hs14741766 28.5 3.8
CE18135 28.5 3.8
7297828_1 28.1 4.2
At2g25530 27.7 5.5
Hs14249610 27.7 5.5
Hs4506231 27.7 5.9
7291779 27.3 8.4
> Hs5803046
Length=334
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 80/129 (62%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A++V + +GWYHSHP YGCWLSGIDV TQ L QQ Q+PF+A+V+DP RT+ +
Sbjct 122 ENAKQVGRLENAIGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKV 181
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT P K D G +E Q +P K+ DFG H K+YY L V + +S+ L
Sbjct 182 NLGAFRT-YPKGYKPPDEGPSEY---QTIPLNKIEDFGVHCKQYYALEVSYFKSSLDRKL 237
Query 121 LEQMCSASW 129
LE + + W
Sbjct 238 LELLWNKYW 246
> 7300154
Length=327
Score = 124 bits (311), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 82/129 (63%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
EAA++V + VGWYHSHP YGCWLSGIDV TQ L Q +Q+PF+AIVVDP+RTV +
Sbjct 119 EAAKEVGRMEHAVGWYHSHPGYGCWLSGIDVSTQMLNQTYQEPFVAIVVDPVRTVSAGKV 178
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT G + ++ E + Q +P K+ DFG H K+YY L + + +++ L
Sbjct 179 CLGAFRTYPKGYKPPNE----EPSEYQTIPLNKIEDFGVHCKQYYPLEISYFKSALDRRL 234
Query 121 LEQMCSASW 129
L+ + + W
Sbjct 235 LDSLWNKYW 243
> At1g71230
Length=358
Score = 123 bits (308), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 55/102 (53%), Positives = 69/102 (67%), Gaps = 4/102 (3%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP YGCWLSGIDV TQ+L QQHQ+PFLA+V+DP RTV +E+G FRT
Sbjct 135 ENVVGWYHSHPGYGCWLSGIDVSTQRLNQQHQEPFLAVVIDPTRTVSAGKVEIGAFRTYS 194
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHW 111
G + D+ + Q +P K+ DFG H K+YY L V +
Sbjct 195 KGYKPPDEP----VSEYQTIPLNKIEDFGVHCKQYYSLDVTY 232
> At1g22920
Length=357
Score = 123 bits (308), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 54/102 (52%), Positives = 68/102 (66%), Gaps = 4/102 (3%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP YGCWLSGIDV TQ L QQ+Q+PFLA+V+DP RTV +E+G FRT
Sbjct 135 ENVVGWYHSHPGYGCWLSGIDVSTQMLNQQYQEPFLAVVIDPTRTVSAGKVEIGAFRTYP 194
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHW 111
G + DD + Q +P K+ DFG H K+YY L + +
Sbjct 195 EGHKISDD----HVSEYQTIPLNKIEDFGVHCKQYYSLDITY 232
> CE06722
Length=368
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/121 (46%), Positives = 78/121 (64%), Gaps = 4/121 (3%)
Query 9 QQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTL 68
++KVVGWYHSHP YGCWLSGIDV TQ L Q+ Q+P++AIV+DP+RT+ +++G FRT
Sbjct 131 KEKVVGWYHSHPGYGCWLSGIDVSTQTLNQKFQEPWVAIVIDPLRTMSAGKVDIGAFRTY 190
Query 69 LPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQMCSAS 128
G DD + Q +P K+ DFG H KRYY L V + + + A +L + ++
Sbjct 191 PEGYRPPDDV----PSEYQSIPLAKIEDFGVHCKRYYSLDVSFFKSQLDAHILTSLWNSY 246
Query 129 W 129
W
Sbjct 247 W 247
> SPAC1687.13c
Length=299
Score = 104 bits (260), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 49/128 (38%), Positives = 78/128 (60%), Gaps = 2/128 (1%)
Query 6 VPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGF 65
V + V+GWYHSHPNYGCWLSG+DV TQ+ Q++QDPF+A+V+DP R++ + + +G F
Sbjct 107 VYRHENVIGWYHSHPNYGCWLSGVDVETQRQNQKYQDPFVAVVLDPKRSLESPYVNIGAF 166
Query 66 RTLLPGEEK--RDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQ 123
RT G + R + + + LP+ K+ D GAH + YY L + + + + E
Sbjct 167 RTYPVGNDGSIRTKSRHHPSVLFKNLPSSKIEDAGAHAEAYYSLPITYFHSKAEKKVTEF 226
Query 124 MCSASWPQ 131
+ + +W +
Sbjct 227 LRNRNWSR 234
> Hs17463759
Length=254
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/131 (37%), Positives = 71/131 (54%), Gaps = 6/131 (4%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A+KV + +GWY+ HP +GCWLSGI TQ L ++ ++PF A+V+DP RT+ +
Sbjct 86 ENAKKVGRLKNAIGWYYRHPGHGCWLSGI--ITQLLNKRFREPFTAVVIDPTRTISAGKV 143
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
L FRT P K D G +E Q + K+ D K+YY L V + S+ L
Sbjct 144 NLDAFRT-YPKGYKPPDEGPSEY---QTILLNKIEDVFVQCKQYYALEVLYFKFSLGCKL 199
Query 121 LEQMCSASWPQ 131
LE + + W +
Sbjct 200 LELLWNKYWAE 210
> YDL216c
Length=455
Score = 88.6 bits (218), Expect = 3e-18, Method: Composition-based stats.
Identities = 36/66 (54%), Positives = 50/66 (75%), Gaps = 3/66 (4%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
VVGW+HSHP Y CWLS ID+ TQ L Q+ QDP++AIVVDP++++ ++ + +G FRT+
Sbjct 174 VVGWFHSHPGYDCWLSNIDIQTQDLNQRFQDPYVAIVVDPLKSLEDKILRMGAFRTI--- 230
Query 72 EEKRDD 77
E K DD
Sbjct 231 ESKSDD 236
> SPAC31G5.13
Length=308
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/128 (35%), Positives = 68/128 (53%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLS +D+ TQQ +Q +A+VVDPI++V + + + FR
Sbjct 105 EMVVGWYHSHPGFGCWLSSVDINTQQSFEQLTPRAVAVVVDPIQSVKGKVV-IDAFRLIN 163
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
TL+ G+E R + G S Q L ++ G H YY LR+++ + +L
Sbjct 164 PSTLMMGQEPRQTTSNLGHINKPSIQAL----IHGLGRH---YYSLRINYKKTELEEIML 216
Query 122 EQMCSASW 129
+ W
Sbjct 217 LNLHKQPW 224
> 7296942
Length=308
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 69/128 (53%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR +
Sbjct 104 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 162
Query 70 P-----GEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
P G+E R + G + S Q L ++ H YY + +++ N + +L
Sbjct 163 PNMLVLGQEPRQTTSNLGHLQKPSVQAL----IHGLNRH---YYSISINYRKNELEQKML 215
Query 122 EQMCSASW 129
+ SW
Sbjct 216 LNLHKKSW 223
> CE19527
Length=312
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 68/128 (53%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCWLSG+D+ TQQ + D +A+VVDPI++V + + + FRT+
Sbjct 108 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSDRAVAVVVDPIQSVKGKVV-IDAFRTIN 166
Query 70 P-----GEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
P +E R + G + S Q L ++ H YY + + + + + +L
Sbjct 167 PQSMALNQEPRQTTSNLGHLQKPSIQAL----IHGLNRH---YYSIPIAYRTHDLEQKML 219
Query 122 EQMCSASW 129
+ SW
Sbjct 220 LNLNKLSW 227
> At5g23540
Length=308
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + +A+VVDPI++V + + + FR
Sbjct 105 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALNQRAVAVVVDPIQSVKGKVV-IDAFRSIN 163
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
T++ G+E R + G S Q L ++ H YY + +++ N + +L
Sbjct 164 PQTIMLGQEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSIAINYRKNELEEKML 216
Query 122 EQMCSASW 129
+ W
Sbjct 217 LNLHKKKW 224
> Hs5031981
Length=310
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> YFR004w
Length=306
Score = 69.7 bits (169), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 60/121 (49%), Gaps = 2/121 (1%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
Q VVGWYHSHP +GCWLS +DV TQ+ +Q +A+VVDPI++V + + + FR +
Sbjct 102 QMVVGWYHSHPGFGCWLSSVDVNTQKSFEQLNSRAVAVVVDPIQSVKGKVV-IDAFRLID 160
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKR-YYQLRVHWCANSISAPLLEQMCSAS 128
G + +++ LL + R YY L + + + +L +
Sbjct 161 TGALINNLEPRQTTSNTGLLNKANIQALIHGLNRHYYSLNIDYHKTAKETKMLMNLHKEQ 220
Query 129 W 129
W
Sbjct 221 W 221
> CE00707
Length=319
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 65/133 (48%), Gaps = 21/133 (15%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCWLS +DV TQQ + +A+VVDPI++V + + L FR++
Sbjct 104 ENVVGWYHSHPGFGCWLSSVDVNTQQSFEALHPRAVAVVVDPIQSVKGK-VMLDAFRSVN 162
Query 70 P-GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWK------------RYYQLRVHWCANSI 116
P + R A AE P + ++ G K +YY L V + S
Sbjct 163 PLNLQIRPLAPTAE-------PRQTTSNLGHLTKPSLISVVHGLGTKYYSLNVAYRMGSN 215
Query 117 SAPLLEQMCSASW 129
+L + SW
Sbjct 216 EQKMLMCLNKKSW 228
> ECU11g0570
Length=294
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 62/127 (48%), Gaps = 12/127 (9%)
Query 9 QQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTL 68
+ VVGWYHSHP +GCWLS +D+ TQQ ++ +A+VVDPI++V + + + FR +
Sbjct 95 HETVVGWYHSHPGFGCWLSTVDISTQQSFEKLCKRAVAVVVDPIQSVKGKVV-IDAFRLI 153
Query 69 ------LPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLE 122
L GE ++ + + L+ ++ H YY + N +L
Sbjct 154 DNQLGVLGGEPRQVTSNIGYLKTPTLISI--IHGLNKH---YYSFNITCRKNDFEQKMLL 208
Query 123 QMCSASW 129
+ +W
Sbjct 209 NLHRKTW 215
> Hs13236583
Length=316
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 3 AEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
AE +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 108 AELTGRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQGFVGLI 153
> At1g10600
Length=271
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
VGW H+HP+ GC++S +D+ T Q AIVV P + S G F+ PG
Sbjct 123 VGWIHTHPSQGCFMSSVDLHTHYSYQVMVPEAFAIVVAPT----DSSKSYGIFKLTDPG 177
> At1g48790
Length=505
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 29/59 (49%), Gaps = 4/59 (6%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
+GW H+HP C++S IDV T Q +AIV+ P + N I FR PG
Sbjct 405 LGWIHTHPTQSCFMSSIDVHTHYSYQIMLPEAVAIVMAPQDSSRNHGI----FRLTTPG 459
> At3g06820
Length=374
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 11 KVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
+V+GWYHSHP+ S +DV TQ + Q F+ ++
Sbjct 87 RVIGWYHSHPHITVLPSHVDVRTQAMYQLLDSGFIGLI 124
> SPAC19B12.10
Length=435
Score = 34.3 bits (77), Expect = 0.065, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 28/58 (48%), Gaps = 6/58 (10%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLP 70
+GW H+HP C++S +D+ T Q +AIV+ P + G FR L P
Sbjct 337 LGWIHTHPTQTCFMSSVDLHTHCSYQLMLPEAIAIVMAPSKNTS------GIFRLLDP 388
> 7301945
Length=420
Score = 33.5 bits (75), Expect = 0.097, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 8 MQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
MQ +GW H+HP +LS +D+ T Q LAIV P
Sbjct 319 MQLITLGWIHTHPTQTAFLSSVDLHTHCSYQIMMPEALAIVCAP 362
> ECU04g0420
Length=258
Score = 32.7 bits (73), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 5 KVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
KV + KV+GWYH+ P + + +D+ T+ L + PFLAI+
Sbjct 71 KVNHKLKVMGWYHTGPR--MYENDLDI-TRSLGSFVESPFLAII 111
> Hs16158201_2
Length=256
Score = 31.2 bits (69), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
V+GWYHSHP + S D+ TQ Q + F+ ++V P
Sbjct 79 VIGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAKFIGMIVSP 122
> Hs5453545
Length=424
Score = 30.8 bits (68), Expect = 0.71, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 331 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESVAIVCSP 369
> YOR261c
Length=338
Score = 30.4 bits (67), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 6/50 (12%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQH--QDPFLAIV 48
E +K+ ++K++GWYHS P L D+ +L +++ +P L IV
Sbjct 78 EMCKKINAKEKLIGWYHSGPK----LRASDLKINELFKKYTQNNPLLLIV 123
> Hs14783322
Length=461
Score = 30.4 bits (67), Expect = 0.94, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 343 LGWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSP 381
> Hs18600963
Length=587
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 1/54 (1%)
Query 6 VPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRS 59
+P + G +HS+P GC + + + TQQ Q Q PF + V RS
Sbjct 331 LPNRLPANGSFHSYPT-GCQPTAVSILTQQAASQWQFPFSSRRVKRTTVTTRRS 383
> Hs18129646
Length=292
Score = 29.6 bits (65), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 10/50 (20%)
Query 48 VVDPIRTVCNRSIELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDF 97
VV+P+RT R LP E K++D+ DA L A ++DF
Sbjct 146 VVEPLRTSVPR----------LPSESKKEDSSDATQVPQASLKASDLSDF 185
> At4g16310
Length=1265
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 7/18 (38%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 7 PMQQKVVGWYHSHPNYGC 24
P++++V+ W+ +H YGC
Sbjct 788 PLERRVMNWHFAHTEYGC 805
> Hs14741766
Length=261
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 28/59 (47%), Gaps = 12/59 (20%)
Query 14 GWYHSHPNYGCWL-SGIDV----------CTQQLQQQHQDPFLAIVVDPIRTVCNRSIE 61
GW S P YG W G+ V TQ++Q++ Q F ++ DP R C+ SI
Sbjct 25 GWKDSTPAYGHWFREGVSVDQETPVATNNSTQKVQKETQGRF-HLLGDPSRNNCSLSIR 82
> CE18135
Length=362
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 5/45 (11%)
Query 5 KVPMQQKVVGWYHSHPNYGCWLSGIDVC-TQQLQQQHQDPFLAIV 48
KV ++K+VGWYH+ P L D+ +QL++ +P L I+
Sbjct 117 KVAAKEKIVGWYHTGPK----LHKNDIAINEQLKRFCPNPVLVII 157
> 7297828_1
Length=567
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Query 9 QQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQ 39
Q +VGWYHSHP + + D C QL Q
Sbjct 353 QLLLVGWYHSHPKFQAEPTLRD-CDAQLDYQ 382
> At2g25530
Length=655
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 24/48 (50%), Gaps = 4/48 (8%)
Query 2 AAEKVPMQQKVVGWYHSHP---NYGCWLSGIDVCTQQLQQQHQDPFLA 46
AEK PMQ + W + P WL+G + QQLQ +H P +A
Sbjct 296 GAEK-PMQYSISSWIMNLPVDEKVKEWLAGEEFYKQQLQMKHILPAVA 342
> Hs14249610
Length=451
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 40/88 (45%), Gaps = 13/88 (14%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQ---QLQQQHQD----PFLAIVVDPIRTVCNRSIE--L 62
+VGWYHSHP+ S D+ Q QL+ Q P LA++ P + N E +
Sbjct 294 LVGWYHSHPHSPALPSLQDIDAQMDYQLRLQGSSNGFQPCLALLCSPYYS-GNPGPESKI 352
Query 63 GGFRTLLPGEEKRDDAG---DAEAASSQ 87
F + P E++ D G D E A Q
Sbjct 353 SPFWVMPPPEQRPSDYGIPMDVEMAYVQ 380
> Hs4506231
Length=324
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 12/47 (25%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query 4 EKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVD 50
+KV ++++VGWYH+ P L D+ +L +++ + +++D
Sbjct 83 KKVNARERIVGWYHTGPK----LHKNDIAINELMKRYCPNSVLVIID 125
> 7291779
Length=338
Score = 27.3 bits (59), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query 4 EKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVD 50
+KV +++VVGWYH+ P L D+ +L +++ + +++D
Sbjct 86 KKVNARERVVGWYHTGPK----LHQNDIAINELVRRYCPNSVLVIID 128
Lambda K H
0.319 0.133 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40