bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2116_orf3
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
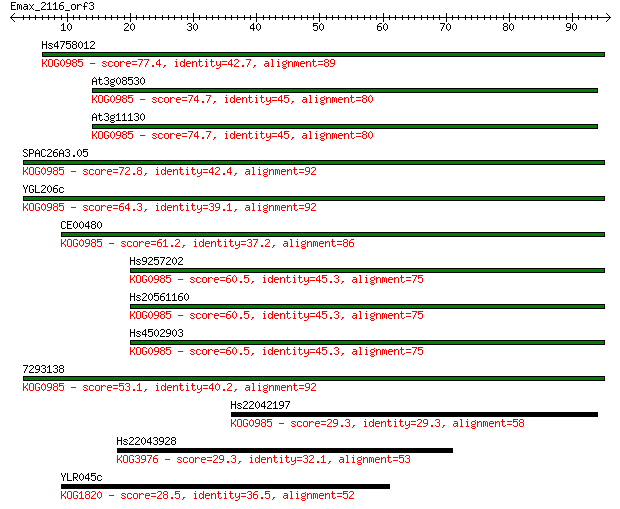
Hs4758012 77.4 5e-15
At3g08530 74.7 4e-14
At3g11130 74.7 4e-14
SPAC26A3.05 72.8 1e-13
YGL206c 64.3 5e-11
CE00480 61.2 5e-10
Hs9257202 60.5 7e-10
Hs20561160 60.5 8e-10
Hs4502903 60.5 8e-10
7293138 53.1 1e-07
Hs22042197 29.3 1.7
Hs22043928 29.3 2.0
YLR045c 28.5 3.3
> Hs4758012
Length=1675
Score = 77.4 bits (189), Expect = 5e-15, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 57/92 (61%), Gaps = 3/92 (3%)
Query 6 QHALPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
++ +P++ N Q + L +A R L GA++ + FN FA G+Y AA++ A G
Sbjct 332 ENIIPYITNVLQNPDLALRMAVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRTP +++F+ VPAQPGQTS +L YF LL+
Sbjct 392 LRTPDTIRRFQSVPAQPGQTSPLLQYFGILLD 423
> At3g08530
Length=1516
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 0/80 (0%)
Query 14 NRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFK 73
N ++ ++LAKR LPGA++ +VQ F + FA Y+ AA + A G LRTP + +F+
Sbjct 166 NNLELAVNLAKRGNLPGAENLVVQRFQELFAQTKYKEAAELAAESPQGILRTPDTVAKFQ 225
Query 74 GVPAQPGQTSAILLYFSTLL 93
VP Q GQT +L YF TLL
Sbjct 226 SVPVQAGQTPPLLQYFGTLL 245
> At3g11130
Length=1705
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 0/80 (0%)
Query 14 NRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFK 73
N ++ ++LAKR LPGA++ +VQ F + FA Y+ AA + A G LRTP + +F+
Sbjct 356 NNLELAVNLAKRGNLPGAENLVVQRFQELFAQTKYKEAAELAAESPQGILRTPDTVAKFQ 415
Query 74 GVPAQPGQTSAILLYFSTLL 93
VP Q GQT +L YF TLL
Sbjct 416 SVPVQAGQTPPLLQYFGTLL 435
> SPAC26A3.05
Length=1666
Score = 72.8 bits (177), Expect = 1e-13, Method: Composition-based stats.
Identities = 39/92 (42%), Positives = 53/92 (57%), Gaps = 4/92 (4%)
Query 3 PYMQHALPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
PY+ L N + + +A LPGAD+ +Q F Q A G+Y AA++ A+ G
Sbjct 334 PYILSNL----NDPGLAVRMASHANLPGADNLYMQQFQQLMAQGNYSEAAKVAASSPRGI 389
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRT QV+ QFK + A PGQ + IL YF TLL+
Sbjct 390 LRTSQVIDQFKLIQAAPGQIAPILQYFGTLLD 421
> YGL206c
Length=1653
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 36/92 (39%), Positives = 51/92 (55%), Gaps = 6/92 (6%)
Query 3 PYMQHALPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
PY+ + L ++ + L +A R GLPGADD + F DY+ AA++ A+ S
Sbjct 342 PYILNKLSNVA----LALIVATRGGLPGADDLFQKQFESLLLQNDYQNAAKVAAS--STS 395
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LR + + K + A PG S ILLYFSTLL+
Sbjct 396 LRNQNTINRLKNIQAPPGAISPILLYFSTLLD 427
> CE00480
Length=1681
Score = 61.2 bits (147), Expect = 5e-10, Method: Composition-based stats.
Identities = 32/89 (35%), Positives = 52/89 (58%), Gaps = 3/89 (3%)
Query 9 LPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRT 65
+P + N+ Q + L LA R LPGA++ V+ FN F++G + +A++ A+ G LRT
Sbjct 333 VPFVTNQLQNPDLALKLAVRCDLPGAEELFVRKFNLLFSNGQFGESAKVAASAPQGILRT 392
Query 66 PQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
P +Q+F+ P+ S +L YF LL+
Sbjct 393 PATIQKFQQCPSTGPGPSPLLQYFGILLD 421
> Hs9257202
Length=1626
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L LA R L GA+ V+ FN FA G Y AA++ A+ G LRT + +Q+F+ +PAQ
Sbjct 349 LRLAVRSNLAGAEKLFVRKFNTLFAQGSYAEAAKVAASAPKGILRTRETVQKFQSIPAQS 408
Query 80 GQTSAILLYFSTLLE 94
GQ S +L YF LL+
Sbjct 409 GQASPLLQYFGILLD 423
> Hs20561160
Length=1285
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L LA R L GA+ V+ FN FA G Y AA++ A+ G LRT + +Q+F+ +PAQ
Sbjct 349 LRLAVRSNLAGAEKLFVRKFNTLFAQGSYAEAAKVAASAPKGILRTRETVQKFQSIPAQS 408
Query 80 GQTSAILLYFSTLLE 94
GQ S +L YF LL+
Sbjct 409 GQASPLLQYFGILLD 423
> Hs4502903
Length=1569
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L LA R L GA+ V+ FN FA G Y AA++ A+ G LRT + +Q+F+ +PAQ
Sbjct 349 LRLAVRSNLAGAEKLFVRKFNTLFAQGSYAEAAKVAASAPKGILRTRETVQKFQSIPAQS 408
Query 80 GQTSAILLYFSTLLE 94
GQ S +L YF LL+
Sbjct 409 GQASPLLQYFGILLD 423
> 7293138
Length=1678
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 37/93 (39%), Positives = 52/93 (55%), Gaps = 5/93 (5%)
Query 3 PYMQHALPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
PY+ L + P+ A L +A R L GA+D V+ FN+ F +G Y AA++ A
Sbjct 336 PYINTVLQN-PDLA---LRMAVRNNLAGAEDLFVRKFNKLFTAGQYAEAAKVAALAPKAI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILL-YFSTLLE 94
LRTPQ +Q+F+ V G T+ LL YF LL+
Sbjct 392 LRTPQTIQRFQQVQTPAGSTTPPLLQYFGILLD 424
> Hs22042197
Length=586
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 36 VQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQPGQTSAILLYFSTLL 93
++ FN+ + G+Y AA A LR + FK V G+ +LL+F L
Sbjct 298 IERFNELISLGEYEKAACYAANSPRRILRNIGTMNTFKAVGKIRGKPLPLLLFFEALF 355
> Hs22043928
Length=164
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 2/53 (3%)
Query 18 IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQ 70
++L +A GL D+L A +H G A+ +LK L +P VLQ
Sbjct 7 VHLDMASSLGL--LTDSLAWALTEHVVPGALSSASTEALSLKKAALPSPGVLQ 57
> YLR045c
Length=888
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 9 LPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGD-YRGAARICATLKS 60
LPH N I G+ D L FN FAS + Y+ AA + +TLK+
Sbjct 819 LPHRVNSLNIRPYRKNGTGVSSVSDDLDIDFNDSFASEESYKRAAAVTSTLKA 871
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40