bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2024_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
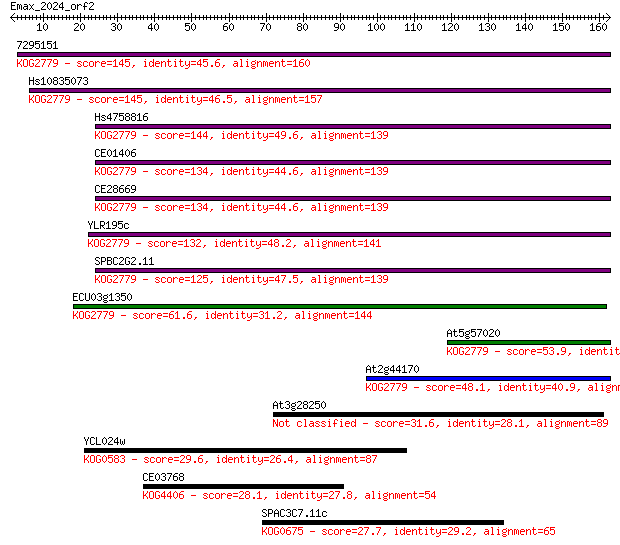
7295151 145 4e-35
Hs10835073 145 4e-35
Hs4758816 144 8e-35
CE01406 134 9e-32
CE28669 134 1e-31
YLR195c 132 3e-31
SPBC2G2.11 125 4e-29
ECU03g1350 61.6 6e-10
At5g57020 53.9 1e-07
At2g44170 48.1 7e-06
At3g28250 31.6 0.70
YCL024w 29.6 2.7
CE03768 28.1 8.1
SPAC3C7.11c 27.7 9.1
> 7295151
Length=472
Score = 145 bits (366), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 73/160 (45%), Positives = 99/160 (61%), Gaps = 4/160 (2%)
Query 3 SLVQAIRGLSLSRSPLNSPHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYN 62
+L+QA+ S + FW TQPV K EQ +TT + I+ K + +++ PY
Sbjct 72 ALLQAVSDAMASTRQMAKKFAFWSTQPVTKLDEQ--VTTNE--CIEPNKEISEIRALPYT 127
Query 63 LPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWI 122
LP GF W + D L ELY LL +YVEDDD +FRF+Y +FL W+L PP + ++W
Sbjct 128 LPGGFKWVTLDLNDANDLKELYTLLNENYVEDDDAMFRFDYQPEFLKWSLQPPGWKRDWH 187
Query 123 IGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+GVRVE S KLVGFISAIP+ L K ++V ++NFLCVH
Sbjct 188 VGVRVEKSGKLVGFISAIPSKLKSYDKVLKVVDINFLCVH 227
> Hs10835073
Length=496
Score = 145 bits (366), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 73/164 (44%), Positives = 99/164 (60%), Gaps = 13/164 (7%)
Query 6 QAIRGLSLSRSPLNS-------PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKK 58
+AI S+ + P + + FWDTQPV K E GP++ K +++++
Sbjct 94 KAIELFSVGQGPAKTMEEASKRSYQFWDTQPVPKLGE----VVNTHGPVEPDK--DNIRQ 147
Query 59 EPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
EPY LP GF W + D VL ELY LL +YVEDDDN+FRF+Y +FL+WAL PP +
Sbjct 148 EPYTLPQGFTWDALDLGDRGVLKELYTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWL 207
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+W GVRV +S+KLVGFISAIP + ++ E+NFLCVH
Sbjct 208 PQWHCGVRVVSSRKLVGFISAIPANIHIYDTEKKMVEINFLCVH 251
> Hs4758816
Length=498
Score = 144 bits (363), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 92/139 (66%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K E G I+ K ++V++EPY+LP GF+W + D +VL EL
Sbjct 121 FWDTQPVPKLDE----VITSHGAIEPDK--DNVRQEPYSLPQGFMWDTLDLSDAEVLKEL 174
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL+WAL PP + +W GVRV +++KLVGFISAIP
Sbjct 175 YTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWLLQWHCGVRVSSNKKLVGFISAIPAN 234
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ ++ E+NFLCVH
Sbjct 235 IRIYDSVKKMVEINFLCVH 253
> CE01406
Length=450
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 62/139 (44%), Positives = 88/139 (63%), Gaps = 2/139 (1%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FW TQPV + E ++ I+ ++ V+ EP++LP GF WS + D + L+EL
Sbjct 68 FWSTQPVPQMDE--TVPADVNCAIEENIALDKVRAEPFSLPAGFRWSNVDLSDEEQLNEL 125
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
YNLL +YVEDDD++FRF+Y DFL WAL P F EW GVR +++ +L+ FI A+P T
Sbjct 126 YNLLTRNYVEDDDSMFRFDYSADFLKWALQVPGFRPEWHCGVRADSNNRLLAFIGAVPQT 185
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ K + + E+NFLCVH
Sbjct 186 VRVYDKTVNMVEINFLCVH 204
> CE28669
Length=452
Score = 134 bits (336), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 62/139 (44%), Positives = 88/139 (63%), Gaps = 2/139 (1%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FW TQPV + E ++ I+ ++ V+ EP++LP GF WS + D + L+EL
Sbjct 70 FWSTQPVPQMDE--TVPADVNCAIEENIALDKVRAEPFSLPAGFRWSNVDLSDEEQLNEL 127
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
YNLL +YVEDDD++FRF+Y DFL WAL P F EW GVR +++ +L+ FI A+P T
Sbjct 128 YNLLTRNYVEDDDSMFRFDYSADFLKWALQVPGFRPEWHCGVRADSNNRLLAFIGAVPQT 187
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ K + + E+NFLCVH
Sbjct 188 VRVYDKTVNMVEINFLCVH 206
> YLR195c
Length=455
Score = 132 bits (331), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 68/141 (48%), Positives = 89/141 (63%), Gaps = 4/141 (2%)
Query 22 HLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLD 81
H FW TQPV E+ +EGPID KT ED+ +P L + F W V++ + L+
Sbjct 38 HKFWRTQPVKDFDEK----VVEEGPIDKPKTPEDISDKPLPLLSSFEWCSIDVDNKKQLE 93
Query 82 ELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIP 141
+++ LL +YVED D FRFNY K+F WAL P + K+W IGVRV+ +QKLV FISAIP
Sbjct 94 DVFVLLNENYVEDRDAGFRFNYTKEFFNWALKSPGWKKDWHIGVRVKETQKLVAFISAIP 153
Query 142 TTLMCNSKRIRVAEVNFLCVH 162
TL K++ E+NFLCVH
Sbjct 154 VTLGVRGKQVPSVEINFLCVH 174
> SPBC2G2.11
Length=466
Score = 125 bits (314), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 87/140 (62%), Gaps = 5/140 (3%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FW TQPV K ++ TQ EGPID + V +EPY L F W+ V + L E+
Sbjct 53 FWKTQPVPKFDDE---CTQ-EGPIDPNTDINQVPREPYRLLKEFEWATIDVTNDNELSEV 108
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
+ LL +YVED + RF Y +FL WAL PP + KEW +GVRV++S+KLV FISA+P +
Sbjct 109 HELLTENYVEDATAMLRFAYISEFLRWALMPPGYVKEWHVGVRVKSSRKLVAFISAVPLS 168
Query 144 LMCNSKRI-RVAEVNFLCVH 162
+ K I + AEVNFLC+H
Sbjct 169 IRVRDKIIKKCAEVNFLCIH 188
> ECU03g1350
Length=355
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 66/144 (45%), Gaps = 19/144 (13%)
Query 18 LNSPHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDP 77
+ H FW TQPV + E+ P T ++E K LP+GF + ED
Sbjct 1 MGKIHKFWSTQPVDRNGEEAM-------PSKHTISIEQPK-----LPDGFRF-----EDL 43
Query 78 QVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFI 137
++EL N L +YVED + Y +FL W + + K + I +R+ ++VGFI
Sbjct 44 GCVEELANFLEKNYVEDIYSGHMLRYSVEFLQWMFDGRDGKKRYCIVLRL--CGRMVGFI 101
Query 138 SAIPTTLMCNSKRIRVAEVNFLCV 161
+ KR V VNFLC+
Sbjct 102 FGKEHLVSIRGKRSSVLGVNFLCI 125
> At5g57020
Length=370
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 31/44 (70%), Gaps = 0/44 (0%)
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+ W IGVR + S+KLV FIS +P + + +++AE+NFLCVH
Sbjct 80 ESWHIGVRAKTSKKLVAFISGVPARIRVRDEVVKMAEINFLCVH 123
> At2g44170
Length=389
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 38/86 (44%), Gaps = 20/86 (23%)
Query 97 NLFRFNYGKDFLVWALNPPNFFKE--------------------WIIGVRVEASQKLVGF 136
L + Y K W P FK+ W IGVRV+ S+KL+ F
Sbjct 58 RLLQCQYSKTHKFWETQPVEQFKDIQDTSLPEGPIEPATLVSESWHIGVRVKTSKKLIAF 117
Query 137 ISAIPTTLMCNSKRIRVAEVNFLCVH 162
I PT + + +++A+VN LCVH
Sbjct 118 ICGRPTRIRVRDEVVKMAKVNSLCVH 143
> At3g28250
Length=121
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 45/102 (44%), Gaps = 18/102 (17%)
Query 72 CSVEDPQVL---------DELYNL----LALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
CS+ D Q++ D +YN+ + L+Y + N + N+G LV +P F+
Sbjct 23 CSIVDCQIISTRGACYSPDNIYNMASVVMNLYYQAEGRNFWNCNFGDSGLVAITDPSEFY 82
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLC 160
+ + + V F I + C + + VA +NF+C
Sbjct 83 ----LSLLFHYTIIYVLFFCLI-SNYFCFRQAMEVANMNFVC 119
> YCL024w
Length=1037
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 45/96 (46%), Gaps = 14/96 (14%)
Query 21 PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVED-----VKKEPYNLPNGFVWSECSVE 75
PH ++ A+ ++S+ + + A +++D +K++P P G +E E
Sbjct 712 PHFTRKSKHFTTANNRRSVLS-----LYAKDSIKDLNEFLIKEDPDLPPQGSTDNESRSE 766
Query 76 DPQVLDELYNLLALHYVEDD----DNLFRFNYGKDF 107
DP++ + + + + Y EDD DN+ N DF
Sbjct 767 DPEIAESITDSRNIQYDEDDSKDGDNVNNDNILSDF 802
> CE03768
Length=444
Score = 28.1 bits (61), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 25/54 (46%), Gaps = 2/54 (3%)
Query 37 KSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDELYNLLALH 90
KS +TQ P T + + + + +P F+ S C P ++D+L L H
Sbjct 225 KSFSTQSNRP--ETPPAQPLPTQQFGVPLEFILSHCGGNIPPIVDQLIEYLEAH 276
> SPAC3C7.11c
Length=560
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 32/69 (46%), Gaps = 5/69 (7%)
Query 69 WSECSVEDPQVLDELY--NLLALHYVEDDDNLFRFN--YGKDFLVWALNPPNFFKEWIIG 124
WS + +P+ + E Y + Y DDD+ F YG F +W + P F +G
Sbjct 349 WSPPMIPNPEFIGEWYPRKIPNPDYF-DDDHPSHFGPLYGVGFELWTMQPNIRFSNIYVG 407
Query 125 VRVEASQKL 133
+E +++L
Sbjct 408 HSIEDAERL 416
Lambda K H
0.319 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40